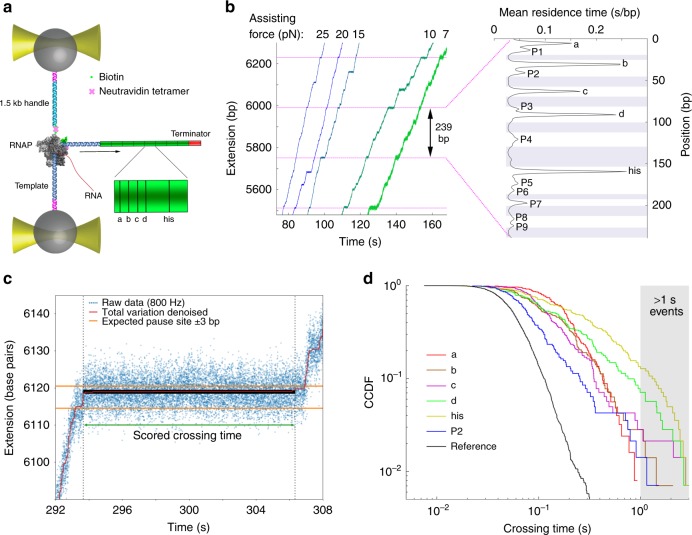
Fig. 1.
Single molecule transcription assay and data analysis. a Experimental geometry. Biotinylated E. coli RNAP halted on the template DNA was tethered through a neutravidin bridge to a biotinylated 1.5 kb DNA ligated to the oligo-coated bead. The selection of which end of the template DNA to ligate to the other beads enables selection between assisting force (illustrated in panel a) and opposing force geometry. The major pause sites (‘his’, ‘a’, ‘b’, ‘c’, ‘d’) are indicated in the sequence of the repeat. b (left) representative traces obtained under assisting forces. Dashed magenta lines were added to highlight the locations of the ‘his’ pause site in the template. The mean residence time histogram shown (right) was calculated by averaging the time spent at each position in the repeat across all traces at all conditions, except RNase data, which was aligned separately (203 traces). Pause sites are marked, and pause-free regions are shaded. For clarity, the mean of measurements up to the 95% percentile is shown to remove to effect of rare pauses occurring outside the pause sites. However, data analysis was performed on the full data set. c Total variation denoising and computation of pause site crossing times. The total variation denoising (red) of the raw data (blue) consists of flat segments separated by discrete jumps. One of these segments occurs in the vicinity of an expected pause site. 1 bp windows are drawn in the ±3 bp range surrounding the expected pause site; the window that took the longest to cross (solid black) is used to define the pause site crossing time for the crossing of this pause site. d Crossing time distributions for different sites measured at 25 pN assisting force. The complementary cumulative distribution function (fraction of events longer than a given crossing time, CCDF) is plotted. The gray shaded area marks the timescales accessible to previous experiments