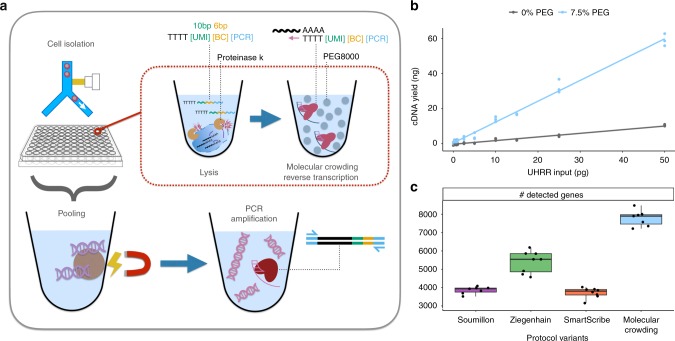
Fig. 1.
mcSCRB-seq workflow and the effect of molecular crowding. a Overview of the mcSCRB-seq protocol workflow. Single cells are isolated via FACS in multiwell plates containing lysis buffer, barcoded oligo-dT primers, and Proteinase K. Reverse transcription and template switching are carried out in the presence of 7.5% PEG 8000 to induce molecular crowding conditions. After pooling the barcoded cDNA with magnetic SPRI beads, PCR amplification using Terra polymerase is performed. b cDNA yield dependent on the absence (gray) or presence (blue) of 7.5% PEG 8000 during reverse transcription and template switching. Shown are three independent reactions for each input concentration of total standardized RNA (UHRR) and the resulting linear model fit. c Number of genes detected (>=1 exonic read) per replicate in RNA-seq libraries, generated from 10 pg of UHRR using four protocol variants (see Supplementary Table 1) at a sequencing depth of one million raw reads. Each dot represents a replicate (n = 8) and each box represents the median and first and third quartiles per method with the whiskers indicating the most extreme data point, which is no more than 1.5 times the length of the box away from the box