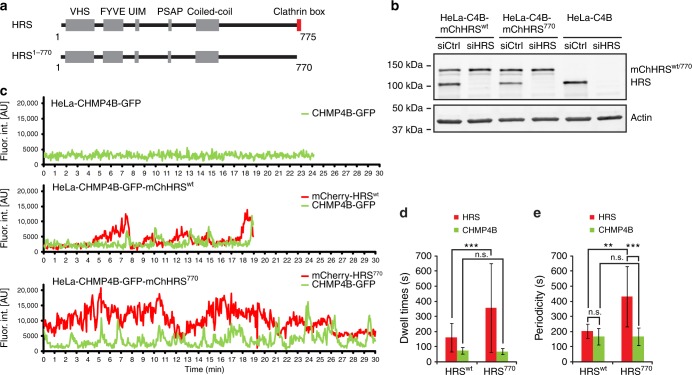
Fig. 5.
Absence of endosomal clathrin leads to hyperstabilization of HRS on endosomes. a Domain structure of HRS and the HRS mutant HRS770 lacking the clathrin-binding domain. b WB showing depletion of endogenous HRS and expression levels of siRNA-resistant mCherry-HRSwt or mCherry-HRS770 in the background of the HeLa-CHMP4B-GFP cell line (“C4B”). c HeLa cells stably expressing CHMP4B-GFP alone or in combination with mCherry-HRSwt or -HRS770 were depleted for endogenous HRS by siRNA as shown in Fig. 5b. Live-cell imaging and fluorescence intensity profiles of tracked EGF-positive endosomes over time. Representative examples of 13 (HRSwt), 12 (HRS770) and 8 (no HRS) tracks from ≥3 independent experiments. d Dwell times for individual mCherry-HRS or CHMP4B-GFP waves were quantified manually in a blinded way. HRSwt: 62 (HRS) and 55 (CHMP4B) waves, HRS770: 29 (HRS770) and 83 (CHMP4B) waves from the dataset described in c. Error bars indicate SD. t-test, ***p < 0.001, n.s. not significant. e Periodicities for mCherry-HRS or CHMP4B-GFP waves were quantified manually in a blinded way. Only tracks covering at least 200 frames were included. HRSwt: 11 tracks, HRS770: 11 tracks from the dataset described in c. Error bars indicate SD. t-test, **p < 0.01, ***p < 0.001, n.s. not statistically significant