Abstract
Background
Probiotics could prevent Pseudomonas aeruginosa colonization in lower respiratory tract (LRT) and reduced P. aeruginosa ventilator-associated pneumonia (VAP) rate. Recent studies also suggested that probiotics could improve lung inflammation in mice infected with P. aeruginosa. It seems that microbiota regulation may be a potential therapy for P. aeruginosa VAP patients. However, we know less about the LRT microbial composition and its correlation with prognosis in P. aeruginosa VAP patients. This study aimed to characterize LRT microbiota in P. aeruginosa VAP patients and explore the relationship between microbiota and patient prognosis.
Methods
Deep endotracheal secretions were sampled from subjects via intubation. Communities were identified by 16S ribosomal RNA gene sequencing. The relationship between microbiota and the prognosis of P. aeruginosa VAP patients were evaluated. Clinical pulmonary infection score and the survival of intensive care unit were both the indicators of patient prognosis.
Results
In this study, the LRT microbial composition of P. aeruginosa VAP patients was significantly different from non-infected intubation patients, and showed significant individual differences, forming two clusters. According to the predominant phylum of each cluster, these two clusters were named Pro cluster and Fir-Bac cluster respectively. Patients from Pro cluster were dominated by Proteobacteria (adj.P < 0.001), while those from Fir-Bac cluster were dominated by Firmicutes, and Bacteroidetes (both adj.P < 0.001). These two varied clusters (Pro and Fir-Bac cluster) were associated with the patients’ primary disease (χ2-test, P < 0.0001). The primary disease of the Pro cluster mainly included gastrointestinal disease (63%), and the Fir-Bac cluster was predominantly respiratory disease (89%). During the two-week dynamic observation period, despite the use of antibiotics, the dominant genera and Shannon diversity of the LRT microbiota did not change significantly in patients with P. aeruginosa VAP. In prognostic analysis, we found a significant negative correlation between Lactobacillus and clinical pulmonary infection score on the day of diagnosis (P = 0.014); but we found no significant difference of microbial composition between survivors and non-survivors.
Conclusions
LRT microbial composition was diversified among P. aeruginosa VAP patients, forming two clusters which were associated with the primary diseases of the patients.
Electronic supplementary material
The online version of this article (10.1186/s12931-018-0847-3) contains supplementary material, which is available to authorized users.
Keywords: Pseudomonas aeruginosa, Mechanical ventilation, Microbiota, Lower respiratory tract, Prognosis, Intensive care unit
Background
Ventilator-associated pneumonia (VAP) is a frequent complication in patients requiring mechanical ventilation and the associated mortality ranges from 20 to 50% [1, 2]. Pseudomonas aeruginosa is one of the most common pathogens causing VAP and is independently associated with increased mortality; in China, it has been staying in the top three pathogens [3–7]. Antibiotic treatment is the primary method for managing P. aeruginosa VAP; however, it constitutes a risk factor for the development of multi-drug resistant P. aeruginosa [2]. Increasing drug resistance, especially in intensive care units (ICUs), could result in P. aeruginosa VAP becoming uncontrollable [8, 9].
Recent findings suggested that the lower respiratory tract (LRT) is inhabited by niche-specific microbiota and VAP occurs mainly when the micro-ecology balance is damaged [10–12]. Probiotics play a preventive role on VAP occurrence, especially the VAP induced by P. aeruginosa [13–15]. Probiotics pre-treated patients obtained a decreased risk of LRT colonization with P. aeruginosa [13, 15]. In addition, Khailova et al. found that Lactobacillus rhamnosus can decrease lung P. aeruginosa load and increase the survival rate of P. aeruginosa pneumoniae mice [16]. It indicated that microbiota regulation could have significant effect on P. aeruginosa infection. However, we know less about the characteristics of LRT microbial composition in P. aeruginosa VAP patients.
In this study, our aim was to examine LRT microbiota characteristics in P. aeruginosa VAP first and then analyze the relationship between LRT microbial characteristics and patient prognosis.
Methods
Subjects
This study was a prospective study conducted at intensive care unit (ICU) of Ruijin Hospital, China. Inclusion criteria of P. aeruginosa VAP patients included [1]: (1) mechanical ventilation > 48 h; (2) satisfied two of the following: body temperature > 38 °C or < 36 °C, leukopenia or leukocytosis, or purulent secretions; (3) new or progressive chest infiltrates, for patients with underlying pulmonary or cardiac disease, two serial chest radiographs were required for assessment; (4) endotracheal aspiration cultured P. aeruginosa at least + 2 growth using semi-quantitative measurements. Exclusion criteria included: (1) age < 18 years; (2) pregnant woman; (3) sputum cultured P. aeruginosa prior to intubation. Clinical data collection was performed at the hospital upon admission and terminated following study withdrawal, discharge, or death. Sequential Organ Failure Assessment (SOFA) score was evaluated at ICU admission and the initial sample collection day. The severity of pulmonary infection in P. aeruginosa VAP patients was assessed using the Clinical Pulmonary Infection Score (CPIS). The criteria of CPIS were performed as previously described [17]. CPIS could be used to assess the clinical outcome of pulmonary infection in patients with VAP [18]. We took the CPIS of the patient at the time of diagnosis as the baseline parameter and reassessed it within 7 days to 14 days after antibiotic treatment; the CPIS score was reduced to within 6 points to be recognized as clinical improvement of pulmonary infection [17]. Control subjects were selected from selective operation patients without acute or chronic respiratory disease, any infection, and antibiotic use for three months prior. Written informed consents were obtained from all study subjects or their lineal consanguinities prior to enrollment. The protocol of this study was approved by the Ruijin Hospital Ethics Committee Shanghai Jiao Tong University School of Medicine.
Sample collection
Initial samples were collected within 24 h post P. aeruginosa VAP diagnosis. Sequential sample collection was performed at day 7 and day 14 post initial sample collection. Endotracheal aspiration samples were collected using an endotracheal tube. Collected samples were stored in sterile 15 mL centrifuge tubes at − 80 °C.
Freeze-drying
We set up negative control using sterile deionized water. Prior to freeze-drying, the sample tubes were transferred into a portable liquid nitrogen container and the sample tubes caps were substituted with parafilm; five small holes were made using sterile yellow tips in a bio-safety cabinet while the sample tubes remained in liquid nitrogen. The sample shelf of the freeze-dryer (FreeZone 6 Liter Console Freeze Dry Systems, Labconco, USA) was pre-cooled for at least 2 h in advance by setting the trap-temperature to − 86 °C. The pre-treated frozen samples were then transferred into the sample shelf in the freeze-dryer (chamber space = 2.82 × 107 mm3, trap-temperature = − 86 °C, and vacuum pressure = 0.165 Torr) for drying. Samples were incubated for 60 h. The samples were hermetically sealed immediately following vacuum release and then stored at − 80 °C.
Bacterial DNA amplification and sequencing
LRT bacterial DNA extraction from freeze-dried powder was performed as previously described [19], including negative controls which set up in the freeze-drying procedure. The V3-V4 region of 16S ribosomal RNA (16S rRNA) gene from each DNA sample was amplified with primers F1 and R2 (5′- CCTACGGGNGGCWGCAG-3′ and 5′-GACTACHVGGGTATCTAATCC-3′) corresponding to positions 341 to 805 of the Escherichia coli 16S rRNA gene using an EasyCycler 96 PCR system (Analytik Jena Corp, AG) with the following program: 3 min at 95 °C (denaturation); 21 cycles of 30 s at 94 °C (denaturation), 30 s at 58 °C (annealing), and 30 s at 72 °C (elongation); 5 min at 72 °C (final extension). The products from different samples were indexed and mixed at equal ratios for sequencing using the Miseq platform (Illumina Inc., USA) according to the manufacturer’s instructions.
Data processing
In this study, negative control samples were not detected any bands when the DNA amplification process was completed. We also sequenced those negative control samples, and a total of 90 sequences were obtained from negative control samples, which can be classified as 31 genera. Within the 31 genera, only two (Escherichia and Streptococcus) belong to the contaminant genera detected in negative controls by previous study [20]. The Escherichia was not found in any particular sample in our study; Streptococcus was the commensal of LRT, and the average relative abundance was 9.7% in the samples from controls and 0.5% in the samples from P. aeruginosa VAP patients.
Raw FASTQ files were demultiplexed and quality-filtered using USEARCH 8.0 with the following criteria: (1) exact index matching, (2) only sequences with > 50 bp overlaps were assembled according to their overlap sequence, (3) merged sequences > 400 bp, and (4) a maximum mismatch in overlap area <0.1. Reads that could not be assembled were discarded. Operational taxonomic units (OTUs) were clustered with 97% similarity cutoff using UPARSE (version 7.1) after chimeric sequences removed. The phylogenetic affiliation of each 16S rRNA gene sequence was analyzed using RDP Classifier against the SILVA (SSU123) 16S rRNA database using a confidence threshold of 70%. The raw sequencing data have been uploaded in the NCBI GenBank Sequence Read Archive database (accession number SRP112361).
Statistical analysis
To access the sequencing depth of the LRT samples, a rarefaction curve was generated as previous study [21] and it indicated that the sequencing depth of samples was reasonable (Additional file 1: Figure S1). Shannon’s diversity was calculated by Mothur. The significant difference in Shannon index between groups was calculated using the Wilcoxon nonparametric test. The unweighted UniFrac distance and weighted UniFrac distance were calculated using Quantitative Insights into Microbial Ecology (QIIME) to assess compositional dissimilarity between samples and finally showed as principal co-ordinates analysis (PCoA) plots conducted in R version 3.2.1 [22]. To find factors that related to the LRT microbial composition in P. aeruginosa VAP patients, patient samples from the initial collection day were subjected to a similarity-based, unsupervised hierarchical clustering [23, 24]. Clustering analysis was performed by unweighted pair-group method with arithmetic means (UPGMA) with Bray Curtis distance [23]. Spearman’s correlation was used to test the relationship between the relative abundance of each genus found in P. aeruginosa VAP patients and CPIS score, SOFA score. The statistic comparison of relative abundance of all taxa between the groups was analyzed using the Wilcoxon rank sum test and the p-values were calculated for the false discovery rate (FDR) (q value) [25]. In the dynamic analysis, the analysis of significant differences in Shannon diversity at different time points using paired nonparametric tests.
Clinical data were analyzed using SPSS version 23 (Armonk, New York, USA). Differences between groups were tested using a two-tailed t-test, Mann-Whitney U test, or chi-squared test as appropriate. Continuous variables were presented as the mean ± standard deviation for normally distributed data and median [interquartile range, IQR] for non-normally distributed data. Categorical variables were presented as number of subjects and percentages. Figures were created using GraphPad Prism version 6.0.
Results
Patient characteristics and sequencing
This study included 36 P. aeruginosa VAP patients and 18 control subjects. Sequential samples were obtained from 26 P. aeruginosa VAP patients for dynamics evaluation. Baseline characteristics are listed in Additional file 1: Table S1. Mean age, which was a known risk factors for VAP, was higher for P. aeruginosa VAP patients than for controls (P = 0.009). All P. aeruginosa VAP patients used antibiotics before mechanical ventilation. There was no significant difference in gender, body mass index (BMI), number of smokers, number of common complications including hypertension and diabetes in P. aeruginosa VAP group and control group (Additional file 1: Table S1).
One sample collected from P. aeruginosa VAP patient in this study failed to sequence, and we have excluded this sample. Finally, there were 94 samples for the further analysis. In total, 2,211,722 sequences were obtained from 94 samples (median 25,391; IQR 13,216–32,270 sequences per sample) by 16S rRNA gene sequencing. Within the 94 samples, 914 OTUs were detected and classified into 19 phyla including 269 genera.
LRT microbiota profiles
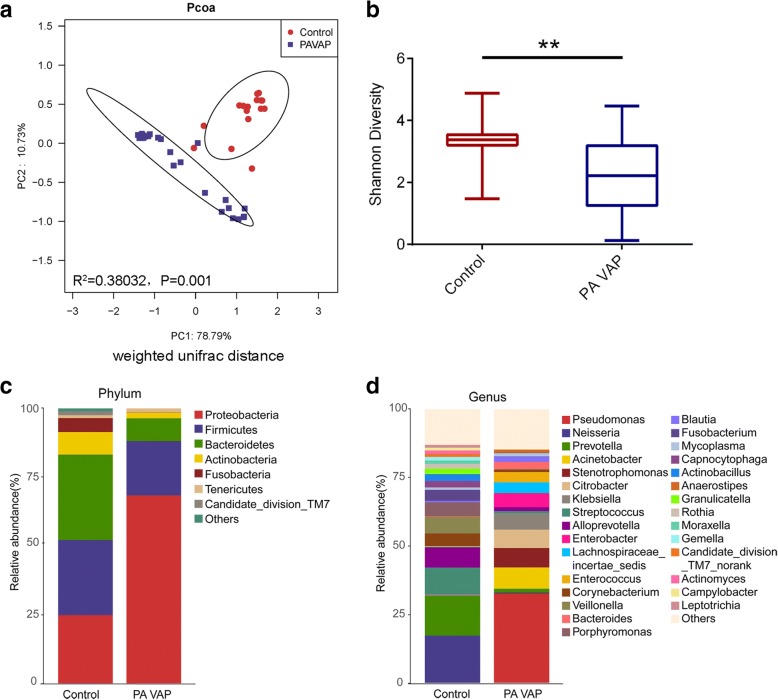
Based on the phylogenetic relationships among the members of the microbial community, the similarity of the LRT microbial composition was measured by the unifac distance. It was found that the composition of the LRT microbiota in patients with P. aeruginosa VAP was significantly different from that in the control group (unweighted unifrac distance, R2 = 0.18431, P = 0.001; weighted unifrac distance, R2 = 0.38032, P = 0.001) (Fig. 1a; Additional file 1: Figure S2). Diversity, presented as Shannon index, was lower in patients with P. aeruginosa VAP compared to the controls (P = 0.0025) (Fig. 1b).
Fig. 1.
The LRT microbiota of P. aeruginosa VAP patients was significantly different from that of the controls. Samples from day1 were collected for analysis. a The weighted unifrac distance between the LRT microbiota of each sample shown as a Pcoa two-dimensional map. Each dot represents one sample. b Shannon diversity index of the P. aeruginosa VAP group was significantly lower than those in the control group. c and d Histogram of lower respiratory tract microbial composition at phylum level (c) and at genus level (d) in P. aeruginosa VAP patients and control subjects. Data were presented as mean ± Standard deviation. The difference between groups was analyzed using the Mann-Whitney test, **P < 0.01. Control, control group; PAVAP, Pseudomonas aeruginosa VAP group
To obtain a global view of LRT microbiota in study subjects, we compared the taxa at phylum and genus level between control and P. aeruginosa VAP group. At phylum level, the dominant taxa of patients in the control group and the P. aeruginosa group were both Proteobacteria, Firmicutes, and Bacteroidetes, but the average abundance of Proteobacteria in the P. aeruginosa group was significantly increased (adj.P = 3.02 × 10− 5) (Fig. 1c). At the genus level, patients in the control group were dominated by Neisseria, Prevotella, Streptococcus and Alloprevotella, while Pseudomonas, Acinetobacter, Enterobacteriaceae (Citrobacter, Klebsiella, Enterobacter) and Lachnospiraceae_incertae_sedis were dominant taxa in the P. aeruginosa group (Fig. 1d).
Primary disease related to LRT microbial clustering of P. aeruginosa VAP patients
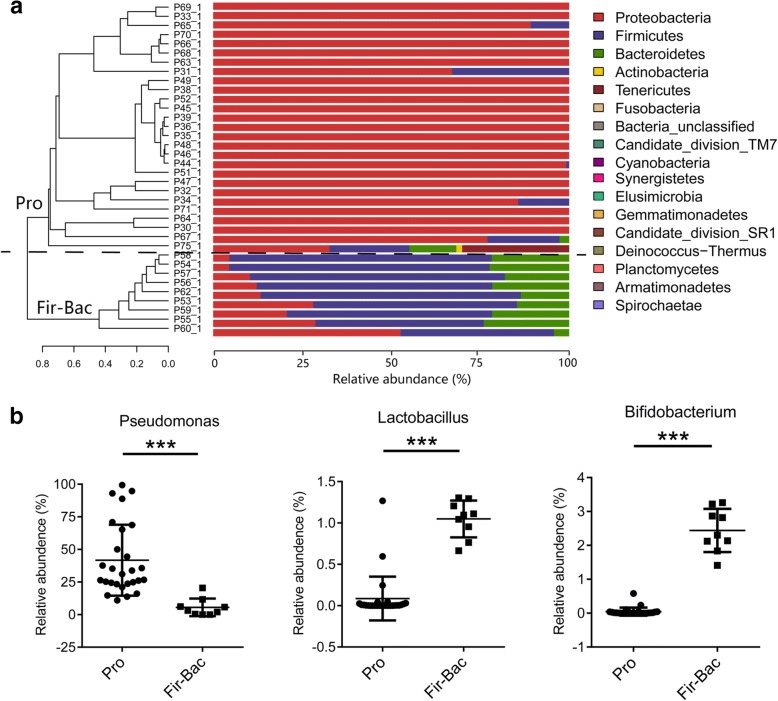
Meanwhile, we found microbial composition within P. aeruginosa VAP group varied; thus, we performed clustering analysis. Samples from P. aeruginosa VAP patients were hierarchically clustered based on similarities in microbial composition (measured by Bray Curtis distance) and visualized as phylum and genus dendrograms with two clusters (Fig. 2a; Additional file 1: Figure S3). Each cluster was given a name according to the most abundant taxon at phylum level and these two clusters were finally termed Pro cluster and Fir-Bac cluster. Pro cluster exhibited a high abundance of Proteobacteria at the phylum level (Fig. 2a) and Pseudomonas, Citrobacter, Enterobacter, Klebsiella, Enterococcus and Acinetobacter at the genus level (Additional file 1: Figure S3). The taxon composition of Fir-Bac cluster consisted mainly of Firmicutes and Bacteroidetes at the phylum level (Fig. 2a) and Lachnospiraceae_incertae_sedis, Bacteroides, Blautia, and Alloprevotella at the genus level (Additional file 1: Figure S3). In addition, we found the abundance of Bifidobacterium and Lactobacillus in Fir-Bac cluster were significantly higher than those of the Pro group (both adj.P < 0.0001), whereas the abundance of Pseudomonas was significantly lower than that of Pro group (adj.P < 0.001) (Fig. 2b). Conventional culture of endotracheal aspiration samples was also obtained from P. aeruginosa VAP patients; the pathogens cultured by conventional methods were accordant with the 16S rRNA sequence data in most of patients (Additional file 1: Table S2, Additional file 1: Figure S3).
Fig. 2.
The LRT microbial composition and hierarchical clustering of P. aeruginosa VAP patients. Initial samples were analyzed. a The Bray curtis distance matrix was used to construct a dendrogram including 36 P. aeruginosa VAP patients. Clusters were separated by black dotted line originating from the top of the dendrogram. The microbial composition at phylum level was visualized as bar charts. Patient number is indicated at the left of each bar. Each bar represents 100% of the OTUs detected per patient; OTUs are color-coded according to phylum. The predominant phylum in Pro cluster was Proteobacteria, and the predominant phylum in Fir-Bac cluster was Firmicutes and Bacteroidetes. b Comparison of different taxa between Pro cluster and Fir-Bac cluster. Data were presented as mean ± Standard deviation. The difference between groups was analyzed using the Mann-Whitney test, ***P < 0.001. LRT lower respiratory tract; VAP, ventilator-associated pneumonia; OTUs, operational taxonomic units
Interestingly, the primary diseases between Pro cluster and Fir-Bac cluster were significantly different. Patients from Pro cluster were mainly suffer from extra-lung disease while patient from Fir-Bac cluster were mainly suffer from lung disease (χ2-test P < 0.0001) (Table 1). Other baseline parameters, including age, body mass index, smoking, chronic coexistence of disease, sepsis, disease severity and the occurrence time of P. aeruginosa VAP, showed no significant difference between Pro cluster and Fir-Bac cluster (Table 1). Disease severity was assessed using Acute Physiology and Chronic Health Evaluation (APACHE) II scores and SOFA scores upon ICU admission.
Table 1.
Baseline characteristics of P. aeruginosa VAP patients classified by LRT microbial similarity
| Characteristics | Pro cluster (n = 27) | Fir-Bac cluster (n = 9) | P value |
|---|---|---|---|
| Demographics | |||
| Age, y | 67.74 ± 14.55 | 69.78 ± 12.44 | 0.709 |
| Gender, female, n (%) | 9 (33%) | 6 (67%) | 0.122 |
| BMI, kg/m2 | 23.45 ± 4.31 | 21.00 ± 4.40 | 0.152 |
| Smoker, n (%) | 6(22%) | 0 (0%) | 0.303 |
| Primary disease | |||
| Respiratory diseasea, n (%) | 2(7%) | 8 (89%) | < 0.0001 |
| Gastrointestinal diseaseb, n (%) | 17 (63%) | 1 (11%) | |
| Neurological disease, n (%) | 4 (15%) | 0 (0%) | |
| Trauma, n (%) | 2 (7%) | 0 (0%) | |
| Bone and joint damage, n (%) | 1 (4%) | 0 (0%) | |
| Autoimmune disease, n (%) | 1 (4%) | 0 (0%) | |
| Chronic coexisting disease | |||
| Chronic bronchitis | 3(11%) | 1(11%) | 1.000 |
| Chronic obstructive pulmonary disease | 2 (7%) | 0(0%) | 1.000 |
| Diabetes, n (%) | 3 (11%) | 3 (33%) | 0.151 |
| Hypertension, n (%) | 12 (44%) | 4 (44%) | 1.000 |
| Disease severity | |||
| APACHE IIc | 18 [16,24] | 16 [9.5–24] | 0.233 |
| SOFAc | 8[2.5,10] | 5[3,7] | 0.263 |
| Diagnosis of sepsis prior to sampling, n (%) | 15 (55%) | 6 (67%) | 0.705 |
| Time of P. aeruginosa VAP onset, dayd | 15 [7–24] | 14 [2–60.5] | 0.971 |
Abbreviations: VAP ventilator-associated pneumonia, BMI body mass index, APACHE II Acute Physiology and Chronic Health Evaluation II, SOFA Sequential Organ Failure Assessment
a. 7 patients with severe pneumonia (not induced by P. aeruginosa), and 1 patient with pharyngeal abscess in Fir-Bac cluster and one with acute exacerbation of chronic obstructive pulmonary disease, and one with hypopharyngeal carcinoma in Pro cluster
b. One patient with acute obstructive suppurative cholangitis in Fir-Bac cluster and 4 with gastrointestinal tumors, 6 with acute pancreatitis, 1 with bile duct stones, 1 with bile duct obstruction, 3 with intestinal obstruction and 2 with gastrointestinal perforation in Pro cluster
c. APACHE II and SOFA scores were calculated within the first 24 h of ICU admission
d. Time of P. aeruginosa VAP onset was defined as the interval from endotracheal intubation to diagnosis of P. aeruginosa VAP
No effect of antibiotics on LRT microbiota in P. aeruginosa VAP patients
Previous studies have suggested that antibiotic was an important factor affecting the microbial composition, so we would explore whether antibiotics are associated with microbial clustering. All patients in this study had been treated with antibiotics prior to sampling in P. aeruginosa VAP patients. Additional file 1: Table S2 showed the use of antibiotics for 36 P. aeruginosa VAP patients one week before sampling. We found that there were no statistically significant differences in aminoglycosides, carbapenems, cephalosporins, fluoroquinolones, and vancomycin in the Pro cluster and the Fir-Bac cluster, and the number of antibiotics used (all P > 0.05) (Additional file 1: Table S4).
The LRT microbiota of P. aeruginosa VAP patients was associated with the severity of VAP
To gain insight into correlation between LRT microbiota and severity of pulmonary infection (measured by CPIS) in patients with P. aeruginosa VAP, we performed Spearman correlation analysis between all genera and CPIS that collected within 24 h post P. aeruginosa VAP diagnosis. It was found that 21 genera showed significantly positive correlation with CPIS while 47 genera showed significantly negative correlation with CPIS (Table 2 and Table 3). Among the genera that showed a significant positive correlation with CPIS in P. aeruginosa VAP patients, Burkholderia, Alcaligenes, Pseudomonas, Massilia, Flavobacterium and Enterobacter can cause lung infections, Sphingomonas is a pathogen that can cause wound infections in animals, and Methylobacterium is often isolated in the hospital environment (all P < 0.05) (Table 2). Lactobacillus and Bifidobacterium were both negatively correlated with CPIS in patients with P. aeruginosa VAP, but there was no statistically significant difference between Bifidobacterium and CPIS (Lactobacillus: R = − 0.4024, P = 0.014; Bifidobacterium: R = − 0.3096, P = 0.066) (Table 3).
Table 2.
Positive correlation between genus of P. aeruginosa VAP and CPIS
| Genus | R | P |
|---|---|---|
| Burkholderia | 0.517183 | 0.001238 |
| Acidaminobacter | 0.483879 | 0.00279 |
| Alcaligenes | 0.441525 | 0.007023 |
| Methylocella | 0.432106 | 0.008495 |
| Sphingomonas | 0.42808 | 0.009199 |
| Acidisoma | 0.426819 | 0.00943 |
| Curvibacter | 0.422908 | 0.010177 |
| Pseudomonas | 0.41731 | 0.011335 |
| Dysgonomonas | 0.400763 | 0.015426 |
| Gemmata | 0.395769 | 0.016881 |
| Macellibacteroides | 0.392997 | 0.017737 |
| Diaphorobacter | 0.39107 | 0.018354 |
| Brevundimonas | 0.383566 | 0.020927 |
| Massilia | 0.378151 | 0.022965 |
| Flavobacterium | 0.37017 | 0.026267 |
| Janthinobacterium | 0.368243 | 0.02712 |
| Methylobacterium | 0.367719 | 0.027356 |
| Petrimonas | 0.366787 | 0.02778 |
| Enterobacter | 0.360325 | 0.030869 |
| Azotobacter | 0.349351 | 0.036758 |
| Reyranella | 0.332149 | 0.047799 |
Table 3.
Negetive correlation between genus of P. aeruginosa VAP and CPIS
| Genus | R | P | Genus | R | P |
|---|---|---|---|---|---|
| Megamonas | −0.584 | 0.00018 | Sutterella | − 0.404 | 0.01465 |
| Anaerovorax | − 0.554 | 0.00046 | Lactobacillus | − 0.402 | 0.01497 |
| Intestinimonas | − 0.511 | 0.00146 | Phascolarctobacterium | −0.394 | 0.01744 |
| Sporosarcina | −0.502 | 0.00180 | Alloprevotella | −0.394 | 0.01750 |
| Thalassospira | −0.495 | 0.00214 | Ruminococcus | −0.392 | 0.01813 |
| Mitsuokella | −0.495 | 0.00214 | Holdemania | −0.391 | 0.01822 |
| Marvinbryantia | −0.495 | 0.00217 | Butyrivibrio | −0.391 | 0.01825 |
| Jeotgalicoccus | −0.494 | 0.00220 | Paraprevotella | −0.384 | 0.02070 |
| Coprobacillus | −0.491 | 0.00234 | Blautia | −0.377 | 0.02330 |
| Howardella | −0.490 | 0.00239 | Erysipelotrichaceae_incertae_sedis | −0.374 | 0.02458 |
| Catenibacterium | −0.482 | 0.00292 | Peptococcus | −0.372 | 0.02530 |
| Coprobacter | −0.480 | 0.00305 | Candidatus_Saccharimonas | −0.370 | 0.02591 |
| Facklamia | −0.466 | 0.00414 | Lachnospiraceae_incertae_sedis | −0.366 | 0.02825 |
| Aeriscardovia | −0.466 | 0.00417 | Ruminococcaceae_incertae_sedis | −0.364 | 0.02904 |
| Dorea | −0.444 | 0.00668 | WCHB1-69_norank | −0.357 | 0.03240 |
| Corynebacterium | −0.441 | 0.00706 | Clostridium_sensu_stricto_1 | −0.355 | 0.03355 |
| Bacteroides | −0.438 | 0.00752 | Acetatifactor | −0.350 | 0.03666 |
| Oscillibacter | −0.432 | 0.00848 | Aerococcus | −0.348 | 0.03727 |
| Atopostipes | −0.431 | 0.00867 | Coprococcus | −0.348 | 0.03759 |
| Roseburia | −0.425 | 0.0098 | Anaerotruncus | −0.342 | 0.04151 |
| Subdoligranulum | −0.421 | 0.01053 | Family_XIII_incertae_sedis | −0.341 | 0.04155 |
| Alistipes | −0.421 | 0.01059 | Megasphaera | −0.339 | 0.04295 |
| Acidaminococcus | −0.409 | 0.01318 | RF9_norank | −0.337 | 0.04455 |
| Anaerostipes | −0.409 | 0.01323 | Bifidobacterium | −0.310 | 0.06610 |
LRT microbiota may not be related to the survival of the ICU in P. aeruginosa VAP patients
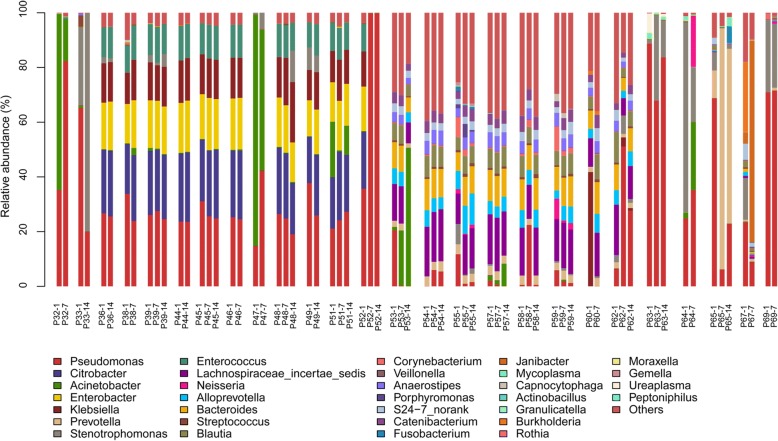
40 sequential samples collected from patients with P. aeruginosa VAP on days 1, 7, and 14 (day 1 was the initial sample collection day). Those patients were all received antibiotics during the two-week observation (Additional file 1: Table S5). The Shannon diversity was relatively stable from day 1 to day 7 and day 1 to day 14 in most P. aeruginosa VAP patients (Additional file 1: Figure S4). The predominant species was stable over time regardless of antibiotic treatment and Pseudomonas proportion fluctuated in nearly all patients (Fig. 3). During the two-week observation, there was no genera changed (paired Wilcoxon test, all adj.P > 0.05).
Fig. 3.
Dynamic variation of LRT microbiota in P. aeruginosa VAP patients two weeks post initial sample collection. Samples of 26 P. aeruginosa VAP patients collected on day 1, day 7, and day 14 were analyzed. Day 1 designates the initial sample collected. Microbial composition is presented as a histogram with patient number and collection time indicated at the bottom of bars. Each bar represents 100% of the OTUs detected per patient and the OTUs are color-coded according to genus. Only genera with a frequency > 5% were included. VAP, ventilator-associated pneumonia; OTUs, operational taxonomic units
During the two-week observation, six patients with P. aeruginosa VAP obtained clinical improvement. No significant taxa was found when we compared the microbial composition between acute phase and clinical improvement phase in those patients (Wilcoxon test of all taxa was adj.P > 0.05). However, of the six patients with P. aeruginosa VAP, abundance of Pseudomonas was decreased in five patients while was increased in one patients (from 21 to 24%) (Additional file 1: Figure S5). We also observed that Lactobacillus was increased in four P. aeruginosa VAP patients and Bifidobacterium was increased in five P. aeruginosa VAP patients (Additional file 1: Figure S5).
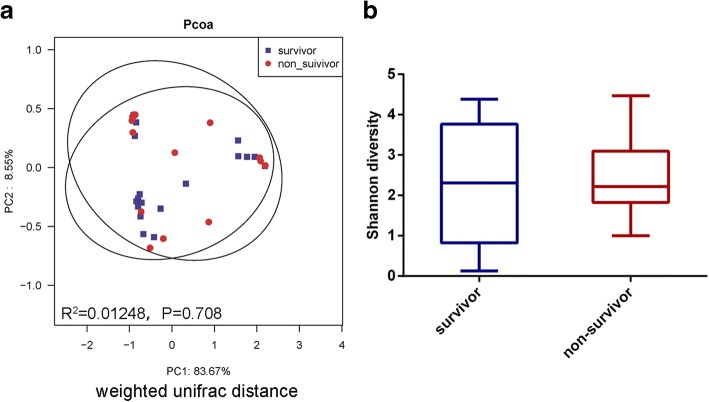
In order to clarify the relationship between LRT microbiota and survival of P. aeruginosa VAP patients, we first performed a Spearman correlation analysis between the relative abundance of genera and SOFA score evaluated in the initial sample collection day. We found that only 4 genera (WCHB1-69_norank, Sphingopyxis, Reyranella and Sphingobacterium) had significant negative correlation with SOFA score (Additional file 1: Table S5). No genus that had significant positive correlation with the SOFA score was found. Next, we divided the P. aeruginosa VAP patients into survival group and non-survival group at the time of discharge. Of the 16 non-survivors, 12 belonged to Pro group and 4 belonged to Fir-Bac group (Pro group vs. Fir-Bac group, χ2-test P > 0.05). The LRT microbial composition and Shannon diversity of the survivors and non-survivors showed no significant difference (unweighted Unifrac distance, R2 = 0.02573, P = 0.468; weighted Unifrac distance, R2 = 0.01248, P = 0.708; Shannon index, P = 0.689) (Fig. 4; Additional file 1: Figure S6).
Fig. 4.
The LRT microbiota of survivors of P. aeruginosa VAP patients had no significant difference compared to that of the non-survivors. Samples from day1 were collected for analysis. a The weighted unifrac distance between the survival group and non-survival group shown as a Pcoa two-dimensional map. Each dot represents one sample. b Shannon diversity index had no statistical difference between survival group and non-survival group (P > 0.05). Data were presented as mean ± Standard deviation. The difference between groups was analyzed using the Mann-Whitney test
Discussion
In this study, we described the dynamic characteristics of the LRT microbiota in patients with P. aeruginosa VAP and their association with clinical characteristics and severity of pneumonia. Microbial composition varied among P. aeruginosa VAP patients, forming two clusters. The microbial clusters related to the primary diseases of P. aeruginosa VAP patients. We also analyzed the correlation between microbial composition and the severity of pneumonia in P. aeruginosa VAP patients. We found that abundance of Pseudomonas was positively correlated with the severity of pneumonia, while the abundance of Lactobacillus was negatively correlated with the severity of pneumonia. .
We found P. aeruginosa VAP patient samples were depleted for LRT commensal bacteria (Streptococcus and Veillonella) and the predominant taxa differed by cluster. Fir-Bac cluster was enriched for Lachnospiraceae_incertae_sedis, Bacteroides, Blautia, and Alloprevotella, and Pro cluster was enriched for Proteobacteria including Enterobacteriaceae and Pseudomonas. Lachnospiraceae_incertae_sedis, Bacteroides, and Blautia are the predominant (and harmless) taxa in normal healthy intestines [26]. Enterobacteriaceae and Pseudomonas are common opportunistic pathogens causing lung infections [2]. Microbial clusters of the LRT microbiota in patients with P. aeruginosa VAP appear to be related to the primary disease. The primary disease in the Pro group was mostly digestive diseases, whereas in the Fir-Bac cluster the patients were mainly respiratory diseases. Previous studies have shown that Lachnospiraceae and Bacteroides are the predominant taxa in fecal samples of patients with respiratory disease such as cystic fibrosis [27], while Enterobacteriaceae increases more frequently in the gut of patients with gastrointestinal diseases [28–30]. Intestinal microbiota can translocate into the lung and that increased intestine permeability is the most probable mechanism underlying microbiota translocation [31]. Few studies have focused on the relationship between LRT microbiota and different primary diseases in patients with hospital infection. There was one study suggested that the predominant taxa in patients with pulmonary sepsis were different from that of the abdominal sepsis [29]. Thus, our findings suggested that the variation in LRT microbiota may be related to the intestinal microbiota of P. aeruginosa VAP patients. However, further analysis of the intestinal microbiota of P. aeruginosa VAP patients is needed.
The CPIS is commonly used to assess the severity of lung infections in patients and the efficacy of antibiotic therapy [17, 18]. In this study, we found that Burkholderia, Pseudomonas, and Enterobacter were positively correlated with CPIS, while Lactobacillus was negatively correlated with CPIS. These results suggested that the LRT microbiota in patients with P. aeruginosa VAP was associated with the severity of pneumonia. We also found that Pseudomonas showed a decreasing trend in the patients that acquired clinical improvement, and Lactobacillus and Bifidobacterium showed an upward trend in those patients. Burkholderia, Pseudomonas, and Enterobacter were common pathogens of hospital-acquired pneumonia [2]. Although they are also present in normal respiratory tract, their abundance is extremely low, and their abundance increases are likely to exacerbate the severity of pneumonia [10, 32]. Lactobacillus and Bifidobacterium could inhibit pathogen proliferation, affect the behavior of other taxa, and regulate the host immune response [33]. Several studies have shown that Lactobacillus and Bifidobacterium have an inhibitory effect on P. aeruginosa, Escherichia coli, Klebsiella pneumonia, and Enterococcus faecalis in vitro [33–36]. Cotar et al. found that Lactobacillus decreases P. aeruginosa virulence by decreasing the expression of lasI, lasR, rhlI, and rhlR involved in quorum sensing and inhibition of biofilm formation [37]. It has also been demonstrated that Bifidobacterium has the ability to inhibit the adhesion of P. aeruginosa to epithelial cells [34]. Therefore, their decline was likely to exacerbate the severity of pneumonia in P. aeruginosa VAP patients. Microbiota regulation may become a potential therapy for P. aeruginosa VAP, however, the issues of delivery and probiotic selection require careful consideration. Although we found LRT microbiota was associated with the severity of pneumonia and early treatment effect, we did not observed difference between survival group and non-survival group. The sampling collection time was far from the death time, ranged from one month to 12 months, which may be one of the reasons that resulted in no significant difference in the composition of the LRT microbial composition between the survivors and non-survivors. On the other hand, we found the SOFA score which was associated with patients’ survival showed little correlation with LRT microbiota in P. aeruginosa VAP patients. It seemed that the LRT microbiota had greater effect on lung than the whole organ system.
Though antibiotic species and numbers were individual in each P. aeruginosa VAP patient, patients of each cluster showed similar microbial composition. Our findings were similar to previous lung-infection-disease studies which showed the lung dominant taxa were stable following the antibiotic treatment [38, 39]. Rogers et al. found that long-term antibiotic therapy did not changed the lung microbial composition in non-cystic fibrosis bronchiectasis patients with baseline airway dominated by P. aeruginosa but changed in those not dominated by P. aeruginosa [40]. The mechanisms were complicated: competitions between bacterial species and antibiotic-bacterial interactions may involve in it. Pseudomonas, Enterobacteriaceae and Acinetobacter were common antibiotic-tolerant pathogens; it was not surprising that they could exist consistently with antibiotic exposure. The consistent microbial dysbiosis had significant negative impact on patient health, so identifying community member interactions and how to break up the network should be pay attention in future studies.
This study had several limitations. First, the sample size was limited. We could not exclude other microbial types that may exist in P. aeruginosa VAP. Second, we characterized the LRT microbiota using the culture-independent technology, which could identify low-proportion or uncultured taxa; however, we were unable to determine the function of specific species accurately. Further analysis regarding probiotic species and associated mechanisms in P. aeruginosa VAP is needed. Third, in this study, the sample collection time was limited. If we could increase the collecting time points in future, it may be better to reflect the changes of the LRT microbiota of patients with P. aeruginosa VAP.
Conclusions
In summary, we described the composition and diversity of the LRT microbiota in patients with P. aeruginosa VAP. Importantly, we have found that in those P. aeruginosa VAP patients, the microbial composition of the LRT is diversified; and that difference was associated with the type of primary disease. We also found that the microbial composition of LRT was related to the severity of P. aeruginosa VAP. Therefore, the LRT microbiota may be a potential target for the treatment of P. aeruginosa VAP.
Additional file
Figure S1. Rarefaction curve of samples collected from P. aeruginosa VAP patients and control subjects. Control, control group; PA VAP, Pseudomonas aeruginosa VAP group. Figure S2. The LRT microbiota was different between P. aeruginosa VAP patients and control subjects. The unweighted unifrac distance between the LRT microbiota of each sample was calculated and it was shown as a Pcoa two-dimensional map. Each dot represents one sample. Control, control group; PAVAP, Pseudomonas aeruginosa VAP group. Figure S3. The LRT microbiota and hierarchical clustering of P. aeruginosa VAP patients at genus level. Initial samples were analyzed. The Bray curtis distance matrix (in genus level) was used to construct a dendrogram including 36 P. aeruginosa VAP patients. Clusters were separated by black dotted line originating from the top of the dendrogram. The microbial composition at genus level was visualized as bar charts. Patient number is indicated at the left of each bar. Each bar represents 100% of the OTUs detected per patient; OTUs are color-coded according to genus. LRT lower respiratory tract; VAP, ventilator-associated pneumonia; OTUs, operational taxonomic units. Figure S4. The Shannon diversity between different time points was stable in LRT samples from P. aeruginosa VAP patients. Comparison of the Shannon diversity between day1 (initial samples) and day7 (A), day1 and day14 (B) were conducted by paired non-parametric test (both P > 0.05). Figure S5. The variety of the relative abundance of Pseudomonas, Lactobacillus and Bifidobacterium in acute phase and improvement phase of P. aeruginosa VAP patients. Pseudomonas (A) showed a decreased trend while Lactobacillus (B) and Bifidobacterium (C) showed an increased trend when the P. aeruginosa VAP patients obtained clinical improvement. Figure S6. The LRT microbiota of survivors of P. aeruginosa VAP patients had no significant difference compared to that of the non-survivors. Samples from day1 were collected for analysis. The unweighted unifrac distance between the survivors and non-survivors was calculated and it was shown as a Pcoa two-dimensional map. Each dot represents one sample. Table S1. Baseline characteristics of control subjects and P. aeruginosa VAP patients. Table S2. Conventional culture of endotracheal aspiration samples from P. aeruginosa VAP patients. Table S3. Antibiotic use before one week sampling of P. aeruginosa VAP patients. Table S4. Antibiotics statistics of P. aeruginosa VAP patients before sampling. Table S5. Antibiotic use during the dynamic observation period of P. aeruginosa VAP patients. Table S6. Negative correlation between genus of P. aeruginosa VAP and SOFA score. (ZIP 1314 kb)
Acknowledgments
Funding
Hongping Qu is supported by National Key R&D Program of China grant (number 2017YFC1309705). Jialin Liu is supported by the Medical-engineering Cross Fund of Shanghai Jiao Tong University grant (number YG2014MS58) and Natural Science Foundation of Shanghai grant (number 16ZR1420900) and National Natural Science Foundation of China grant (number 81770005). Jieming Qu is supported by Shanghai Key discipline for respiratory diseases grant (number 2017ZZ02014).
Availability of data and materials
Not applicable.
Abbreviations
- APACHE
Acute Physiology and Chronic Health Evaluation
- BMI
Body mass index
- CPIS
Clinical Pulmonary Infection Score
- FDR
False discovery rate
- ICU
Intensive care unit
- IQR
Interquartile range
- LRT
lower respiratory tract
- OTU
Operational taxonomic unit
- PCoA
Principal co-ordinates analysis
- SOFA
Sequential Organ Failure Assessment
- UPGMA
Unweighted pair-group method witharithmetic means
- VAP
Ventilator-associated pneumonia
Authors’ contributions
YH, JQ, JL contributed to study design. XQ and HQ contributed to the subject recruitment and the sample collection. DY, LZ, YH contributed to freeze-drying and 16S rRNA gene sequencing. XQ, HQ, YY contributed to clinical data collection and bioinformatics analysis. YH, JQ, JL contributed to data interpretation. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Written informed consents were obtained from all study subjects or their lineal consanguinities prior to enrollment. The protocol of this study was approved by the Ruijin Hospital Ethics Committee Shanghai Jiao Tong University School of Medicine.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Xiaoling Qi and Hongping Qu contributed equally to this work.
Electronic supplementary material
The online version of this article (10.1186/s12931-018-0847-3) contains supplementary material, which is available to authorized users.
Contributor Information
Xiaoling Qi, Email: connie_qi@sjtu.edu.cn.
Hongping Qu, Email: qhp10516@rjh.com.cn.
Dandan Yang, Email: hedyfly@sjtu.edu.cn.
Lian Zhou, Email: lianzhou@sjtu.edu.cn.
Ya-Wen He, Email: yawenhe@sjtu.edu.cn.
Yuetian Yu, Email: fishyyt@sina.com.
Jieming Qu, Email: jmqu0906@163.com.
Jialin Liu, Email: ljl11243@rjh.com.cn.
References
- 1.American Thoracic S Infectious diseases Society of a: guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am J Respir Crit Care Med. 2005;171:388–416. doi: 10.1164/rccm.200405-644ST. [DOI] [PubMed] [Google Scholar]
- 2.Kalil AC, Metersky ML, Klompas M, Muscedere J, Sweeney DA, Palmer LB, Napolitano LM, O'Grady NP, Bartlett JG, Carratala J, et al. Management of Adults With Hospital-acquired and Ventilator-associated Pneumonia: 2016 Clinical practice guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin Infect Dis. 2016;63:e61–e111. doi: 10.1093/cid/ciw353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vincent JL, Sakr Y, Sprung CL, Ranieri VM, Reinhart K, Gerlach H, Moreno R, Carlet J, Le Gall JR, Payen D. Sepsis occurrence in acutely ill patients I: Sepsis in European intensive care units: results of the SOAP study. Crit Care Med. 2006;34:344–353. doi: 10.1097/01.CCM.0000194725.48928.3A. [DOI] [PubMed] [Google Scholar]
- 4.Klompas M, Kleinman K, Murphy MV. Descriptive epidemiology and attributable morbidity of ventilator-associated events. Infect Control Hosp Epidemiol. 2014;35:502–510. doi: 10.1086/675834. [DOI] [PubMed] [Google Scholar]
- 5.Borgatta B, Gattarello S, Mazo CA, Imbiscuso AT, Larrosa MN, Lujan M, Rello J. The clinical significance of pneumonia in patients with respiratory specimens harbouring multidrug-resistant Pseudomonas aeruginosa: a 5-year retrospective study following 5667 patients in four general ICUs. Eur J Clin Microbiol Infect Dis. 2017;36(11):2155–63. [DOI] [PubMed]
- 6.Tao L, Hu B, Rosenthal VD, Gao X, He L. Device-associated infection rates in 398 intensive care units in shanghai, China: international nosocomial infection control consortium (INICC) findings. Int J Infect Dis. 2011;15:e774–e780. doi: 10.1016/j.ijid.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 7.Ding C, Yang Z, Wang J, Liu X, Cao Y, Pan Y, Han L, Zhan S. Prevalence of Pseudomonas aeruginosa and antimicrobial-resistant Pseudomonas aeruginosa in patients with pneumonia in mainland China: a systematic review and meta-analysis. Int J Infect Dis. 2016;49:119–128. doi: 10.1016/j.ijid.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 8.Rosenthal VD, Bijie H, Maki DG, Mehta Y, Apisarnthanarak A, Medeiros EA, Leblebicioglu H, Fisher D, Alvarez-Moreno C, Khader IA, et al. International nosocomial infection control consortium (INICC) report, data summary of 36 countries, for 2004-2009. Am J Infect Control. 2012;40:396–407. doi: 10.1016/j.ajic.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 9.Gupta R, Malik A, Rizvi M, Ahmed M, Singh A. Epidemiology of multidrug-resistant gram-negative pathogens isolated from ventilator-associated pneumonia in ICU patients. J Glob Antimicrob Resist. 2017;9:47–50. doi: 10.1016/j.jgar.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 10.Kelly BJ, Imai I, Bittinger K, Laughlin A, Fuchs BD, Bushman FD, Collman RG. Composition and dynamics of the respiratory tract microbiome in intubated patients. Microbiome. 2016;4:7. doi: 10.1186/s40168-016-0151-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zakharkina T, Martin-Loeches I, Matamoros S, Povoa P, Torres A, Kastelijn JB, Hofstra JJ, de Wever B, de Jong M, Schultz MJ, et al. The dynamics of the pulmonary microbiome during mechanical ventilation in the intensive care unit and the association with occurrence of pneumonia. Thorax. 2017;72:803–810. doi: 10.1136/thoraxjnl-2016-209158. [DOI] [PubMed] [Google Scholar]
- 12.Man WH, de Steenhuijsen Piters WA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol. 2017;15:259–270. doi: 10.1038/nrmicro.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forestier C, Guelon D, Cluytens V, Gillart T, Sirot J, De Champs C. Oral probiotic and prevention of Pseudomonas aeruginosa infections: a randomized, double-blind, placebo-controlled pilot study in intensive care unit patients. Crit Care. 2008;12:R69. doi: 10.1186/cc6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrow LE, Kollef MH, Casale TB. Probiotic prophylaxis of ventilator-associated pneumonia: a blinded, randomized, controlled trial. Am J Respir Crit Care Med. 2010;182:1058–1064. doi: 10.1164/rccm.200912-1853OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siempos II, Ntaidou TK, Falagas ME. Impact of the administration of probiotics on the incidence of ventilator-associated pneumonia: a meta-analysis of randomized controlled trials. Crit Care Med. 2010;38:954–962. doi: 10.1097/CCM.0b013e3181c8fe4b. [DOI] [PubMed] [Google Scholar]
- 16.Khailova L, Baird CH, Rush AA, McNamee EN, Wischmeyer PE. Lactobacillus rhamnosus GG improves outcome in experimental pseudomonas aeruginosa pneumonia: potential role of regulatory T cells. Shock. 2013;40:496–503. doi: 10.1097/SHK.0000000000000066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kollef MH, Ricard JD, Roux D, Francois B, Ischaki E, Rozgonyi Z, Boulain T, Ivanyi Z, Janos G, Garot D, et al. A randomized trial of the amikacin Fosfomycin inhalation system for the adjunctive therapy of gram-negative ventilator-associated pneumonia: IASIS trial. Chest. 2017;151:1239–1246. doi: 10.1016/j.chest.2016.11.026. [DOI] [PubMed] [Google Scholar]
- 18.Luna CM, Blanzaco D, Niederman MS, Matarucco W, Baredes NC, Desmery P, Palizas F, Menga G, Rios F, Apezteguia C. Resolution of ventilator-associated pneumonia: prospective evaluation of the clinical pulmonary infection score as an early clinical predictor of outcome. Crit Care Med. 2003;31:676–682. doi: 10.1097/01.CCM.0000055380.86458.1E. [DOI] [PubMed] [Google Scholar]
- 19.Godon JJ, Zumstein E, Dabert P, Habouzit F, Moletta R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl Environ Microbiol. 1997;63:2802–2813. doi: 10.1128/aem.63.7.2802-2813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Zhao Y, Xu J, Xue Z, Zhang M, Pang X, Zhang X, Zhao L. Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci Rep. 2015;5:14405. doi: 10.1038/srep14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, Panzer AR, LaMere B, Rackaityte E, Lukacs NW, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016;22:1187–1191. doi: 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dogra S, Sakwinska O, Soh SE, Ngom-Bru C, Bruck WM, Berger B, Brussow H, Lee YS, Yap F, Chong YS, et al. Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. MBio. 2015;6 [DOI] [PMC free article] [PubMed]
- 24.de Steenhuijsen Piters WA, Heinonen S, Hasrat R, Bunsow E, Smith B, Suarez-Arrabal MC, Chaussabel D, Cohen DM, Sanders EA, Ramilo O, et al. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med. 2016;194:1104–1115. doi: 10.1164/rccm.201602-0220OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu R, Hong J, Xu X, Feng Q, Zhang D, Gu Y, Shi J, Zhao S, Liu W, Wang X, et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med. 2017;23:859–868. doi: 10.1038/nm.4358. [DOI] [PubMed] [Google Scholar]
- 26.Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte D, et al. Population-level analysis of gut microbiome variation. Science. 2016;352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 27.Burke DG, Fouhy F, Harrison MJ, Rea MC, Cotter PD, O'Sullivan O, Stanton C, Hill C, Shanahan F, Plant BJ, Ross RP. The altered gut microbiota in adults with cystic fibrosis. BMC Microbiol. 2017;17:58. doi: 10.1186/s12866-017-0968-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan C, Ling Z, Huang Y, Cao Y, Liu Q, Cai T, Yuan H, Liu C, Li Y, Xu K. Dysbiosis of intestinal microbiota associated with inflammation involved in the progression of acute pancreatitis. Pancreas. 2015;44:868–875. doi: 10.1097/MPA.0000000000000355. [DOI] [PubMed] [Google Scholar]
- 29.Lankelma JM, van Vught LA, Belzer C, Schultz MJ, van der Poll T, de Vos WM, Wiersinga WJ. Critically ill patients demonstrate large interpersonal variation in intestinal microbiota dysregulation: a pilot study. Intensive Care Med. 2017;43:59–68. doi: 10.1007/s00134-016-4613-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang R, Wei Y, Li Y, Chen W, Chen H, Wang Q, Yang F, Miao Q, Xiao X, Zhang H, et al. Gut microbial profile is altered in primary biliary cholangitis and partially restored after UDCA therapy. Gut. 2018;67:534–541. doi: 10.1136/gutjnl-2016-313332. [DOI] [PubMed] [Google Scholar]
- 31.Klingensmith NJ, Coopersmith CM. The gut as the Motor of Multiple Organ Dysfunction in critical illness. Crit Care Clin. 2016;32:203–212. doi: 10.1016/j.ccc.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, Bushman FD, Collman RG. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bron PA, van Baarlen P, Kleerebezem M. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat Rev Microbiol. 2011;10:66–78. doi: 10.1038/nrmicro2690. [DOI] [PubMed] [Google Scholar]
- 34.Matsumoto T, Ishikawa H, Tateda K, Yaeshima T, Ishibashi N, Yamaguchi K. Oral administration of Bifidobacterium longum prevents gut-derived Pseudomonas aeruginosa sepsis in mice. J Appl Microbiol. 2008;104:672–680. doi: 10.1111/j.1365-2672.2007.03593.x. [DOI] [PubMed] [Google Scholar]
- 35.Rao KP, Chennappa G, Suraj U, Nagaraja H, Raj AP, Sreenivasa MY. Probiotic potential of lactobacillus strains isolated from sorghum-based traditional fermented food. Probiotics Antimicrob Proteins. 2015;7:146–156. doi: 10.1007/s12602-015-9186-6. [DOI] [PubMed] [Google Scholar]
- 36.Shokri D, Khorasgani MR, Mohkam M, Fatemi SM, Ghasemi Y, Taheri-Kafrani A. The inhibition effect of lactobacilli against growth and biofilm formation of Pseudomonas aeruginosa. Probiotics Antimicrob Proteins. 2017;10(1):34–42. [DOI] [PubMed]
- 37.Cotar AI, Saviuc C, Nita RA, Bezirtzoglou E, Lazar V, Chifiriuc MC. Anti-pathogenic strategies for fighting Pseudomonas aeruginosa infections- probiotic soluble compounds as inhibitors of quorum sensing genes expression. Curr Org Chem. 2013;17:155–161. doi: 10.2174/1385272811317020012. [DOI] [Google Scholar]
- 38.Wang Z, Bafadhel M, Haldar K, Spivak A, Mayhew D, Miller BE, Tal-Singer R, Johnston SL, Ramsheh MY, Barer MR, et al. Lung microbiome dynamics in COPD exacerbations. Eur Respir J. 2016;47:1082–1092. doi: 10.1183/13993003.01406-2015. [DOI] [PubMed] [Google Scholar]
- 39.Heirali AA, Workentine ML, Acosta N, Poonja A, Storey DG, Somayaji R, Rabin HR, Whelan FJ, Surette MG, Parkins MD. The effects of inhaled aztreonam on the cystic fibrosis lung microbiome. Microbiome. 2017;5:51. doi: 10.1186/s40168-017-0265-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogers GB, Bruce KD, Martin ML, Burr LD, Serisier DJ. The effect of long-term macrolide treatment on respiratory microbiota composition in non-cystic fibrosis bronchiectasis: an analysis from the randomised, double-blind, placebo-controlled BLESS trial. Lancet Respir Med. 2014;2:988–996. doi: 10.1016/S2213-2600(14)70213-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Rarefaction curve of samples collected from P. aeruginosa VAP patients and control subjects. Control, control group; PA VAP, Pseudomonas aeruginosa VAP group. Figure S2. The LRT microbiota was different between P. aeruginosa VAP patients and control subjects. The unweighted unifrac distance between the LRT microbiota of each sample was calculated and it was shown as a Pcoa two-dimensional map. Each dot represents one sample. Control, control group; PAVAP, Pseudomonas aeruginosa VAP group. Figure S3. The LRT microbiota and hierarchical clustering of P. aeruginosa VAP patients at genus level. Initial samples were analyzed. The Bray curtis distance matrix (in genus level) was used to construct a dendrogram including 36 P. aeruginosa VAP patients. Clusters were separated by black dotted line originating from the top of the dendrogram. The microbial composition at genus level was visualized as bar charts. Patient number is indicated at the left of each bar. Each bar represents 100% of the OTUs detected per patient; OTUs are color-coded according to genus. LRT lower respiratory tract; VAP, ventilator-associated pneumonia; OTUs, operational taxonomic units. Figure S4. The Shannon diversity between different time points was stable in LRT samples from P. aeruginosa VAP patients. Comparison of the Shannon diversity between day1 (initial samples) and day7 (A), day1 and day14 (B) were conducted by paired non-parametric test (both P > 0.05). Figure S5. The variety of the relative abundance of Pseudomonas, Lactobacillus and Bifidobacterium in acute phase and improvement phase of P. aeruginosa VAP patients. Pseudomonas (A) showed a decreased trend while Lactobacillus (B) and Bifidobacterium (C) showed an increased trend when the P. aeruginosa VAP patients obtained clinical improvement. Figure S6. The LRT microbiota of survivors of P. aeruginosa VAP patients had no significant difference compared to that of the non-survivors. Samples from day1 were collected for analysis. The unweighted unifrac distance between the survivors and non-survivors was calculated and it was shown as a Pcoa two-dimensional map. Each dot represents one sample. Table S1. Baseline characteristics of control subjects and P. aeruginosa VAP patients. Table S2. Conventional culture of endotracheal aspiration samples from P. aeruginosa VAP patients. Table S3. Antibiotic use before one week sampling of P. aeruginosa VAP patients. Table S4. Antibiotics statistics of P. aeruginosa VAP patients before sampling. Table S5. Antibiotic use during the dynamic observation period of P. aeruginosa VAP patients. Table S6. Negative correlation between genus of P. aeruginosa VAP and SOFA score. (ZIP 1314 kb)
Data Availability Statement
Not applicable.