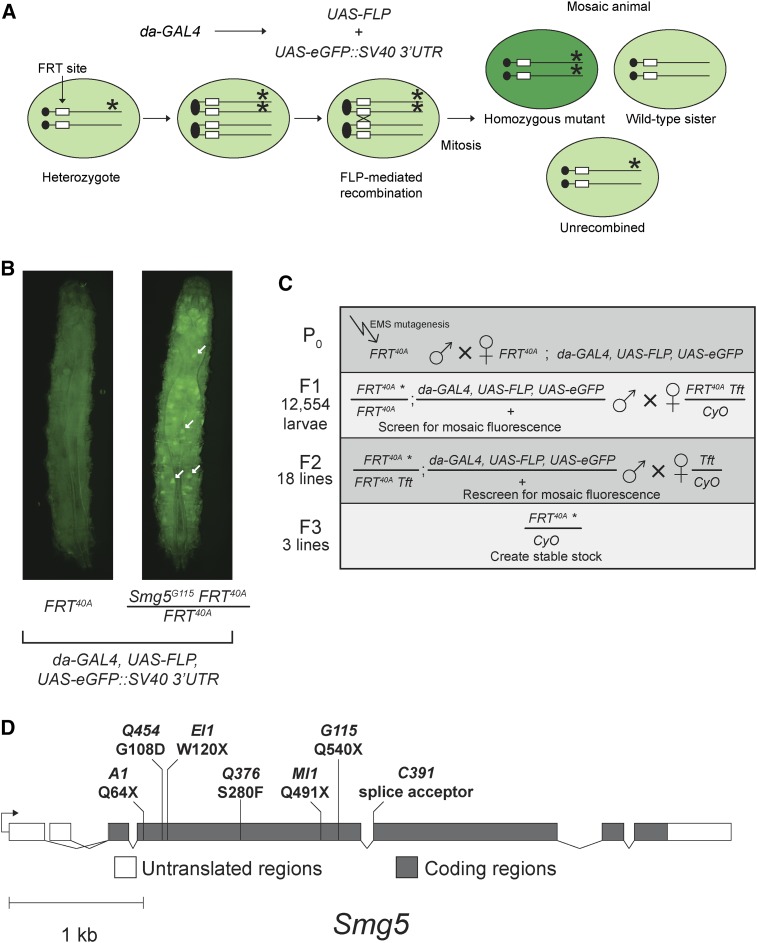
Figure 1.
Mosaic screen for novel NMD-defective mutations on the second chromosome identifies Smg5 alleles. (A) Scheme to generate mosaics and detect mutants with defective NMD. The GAL4 transcription factor is ubiquitously expressed under a daughterless (da) promoter and activates transcription of FLP recombinase and the NMD-sensitive eGFP::SV40 3′UTR fluorescent reporter, both under UAS control. The reporter mRNA is usually degraded by NMD, and thus cells lacking NMD activity due to a homozygous mutation in a gene required for NMD activity show increased green fluorescence. (B) Example of mosaic GFP-reporter fluorescence phenotype detected in our screen. Late L3 larvae expressing the NMD-sensitive eGFP::SV40 3′UTR fluorescent reporter in animals with a wild-type FRT40A chromosome (left) or an FRT40A Smg5G115 chromosome (right). Individual homozygous mutant cells with increased GFP fluorescence in the Smg5 mutant animal are indicated by white arrows. Overall increased fluorescence is due to other out-of-focus mutant cells. Dorsal view; anterior at top. (C) Scheme for recovering mutations identified in the screen. The total number of candidate mutants scored in each generation is shown on the left side of each row. Genotypes on the left in the F1 and F2 generations are the offspring from the previous mating. (D) Molecular identity of isolated Smg5 mutations. Four alleles (A1, EI1, MI1, and G115) are nonsense mutations. C391 is a mutation in a splice acceptor site. Q454 and Q376 are missense mutations. The codon changes for all Smg5 alleles are listed in Table S2.