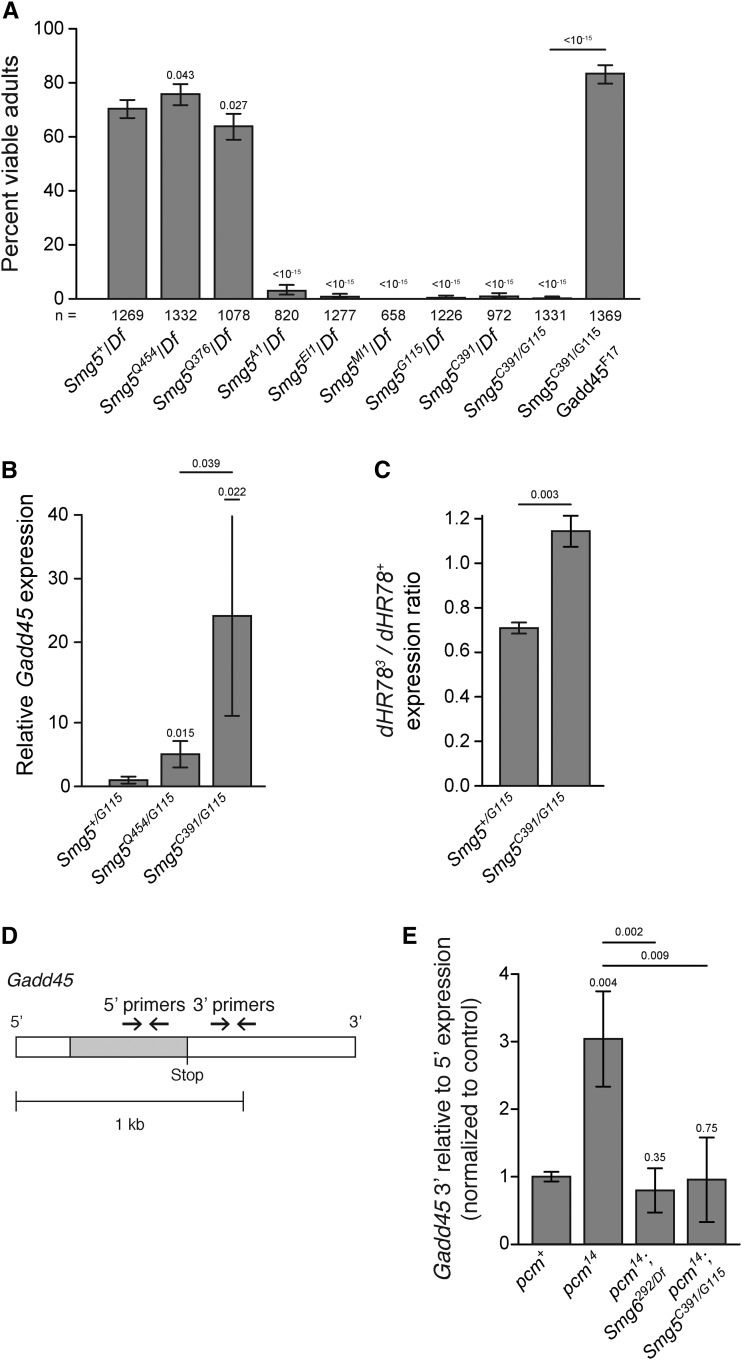
Figure 2.
Smg5 is required for viability and NMD activity in Drosophila. (A) Adult viability of Smg5 mutant alleles trans-heterozygous to either a deficiency removing the Smg5 locus (Df) or other Smg5 mutant alleles. Error bars represent 95% confidence interval of the binomial distribution. P-value listed compared to Smg5+/Df condition or between indicated conditions determined by the test of equal or given proportions. n = total number of animals scored. (B) Expression of the endogenous NMD target Gadd45, as measured by qRT-PCR. Error bars represent 2 SEM. P-value listed for each condition compared to controls or between indicated conditions, determined by two-sided Student’s t-test. n ≥ 3 for all conditions. qPCR primers used were located 5′ to the Gadd45 termination sequence. (C) Relative abundance of PTC-containing dHR783 allele (Fisk and Thummel 1998) mRNA compared to wild-type dHR78 allele mRNA in animals heterozygous for dHR783 in each indicated genotype. Error bars represent 2 SEM. P-value listed for each condition compared to +/Smg5G115, determined by two-sided Student’s t-test. n = 3 for all conditions. (D) Diagram of the endogenous NMD target Gadd45 transcript and 5′ and 3′ qRT-PCR primer pairs. Open boxes indicate UTRs; gray boxes indicate coding regions. 5′ primer pair is located 5′ to the stop codon and the 3′ primer pair is 3′ to the stop codon. (E) Gadd45 expression measured with the 3′ primer pair relative to the 5′ primer pair. The 3′ region is preferentially stabilized in pcm14 mutants. This preferential stabilization is lost when either Smg6 or Smg5 are lost. Error bars represent 2 SEM. P-value listed for each condition compared to the pcm+ condition or between indicated conditions, determined by two-sided Student’s t-test. n ≥ 3 for all conditions.