Figure 1.
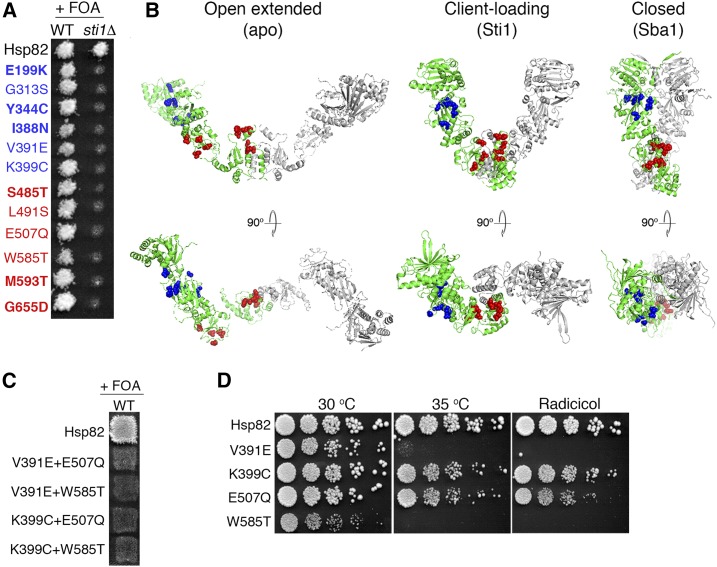
Hsp90 mutations that cause Sti1 dependence cluster in two regions. (A) Plasmid shuffle of indicated Hsp82 mutants in strains MR318 (STI1+, WT) and 1670 (sti1∆). All of the mutants support growth of the STI1+ strain, but none support growth of the sti1∆ strain on FOA, indicating dependence on Sti1. Those indicated in bold were identified in this study. (B) Mutations in (A) are represented as spheres on the various tertiary conformations of Hsp90. For clarity, only one monomer of the Hsp90 homodimer is colored. Left, the open-extended, apo form of Hsc82 (Krukenberg et al. 2009); middle, the Hop-bound conformation of Hsp90 (Southworth and Agard 2011); right, the closed, Sba1-bound form of Hsp82 (Ali et al. 2006). (C) A similar plasmid experiment as in (A) with the indicated double mutants in strain MR318 (WT). Combining SdN and SdC mutations was lethal even in these STI1+ cells. (D) Log-phase cultures of cells recovered from STI1+ cells on FOA plates in (A) that express the indicated Hsp82 mutants as the only source of Hsp90 were serially diluted and plated on rich medium without (left and middle) or with radicicol (right). All Hsp82 mutants have growth defects with V391E and W585T exhibiting the most severe phenotypes. WT, wild type.