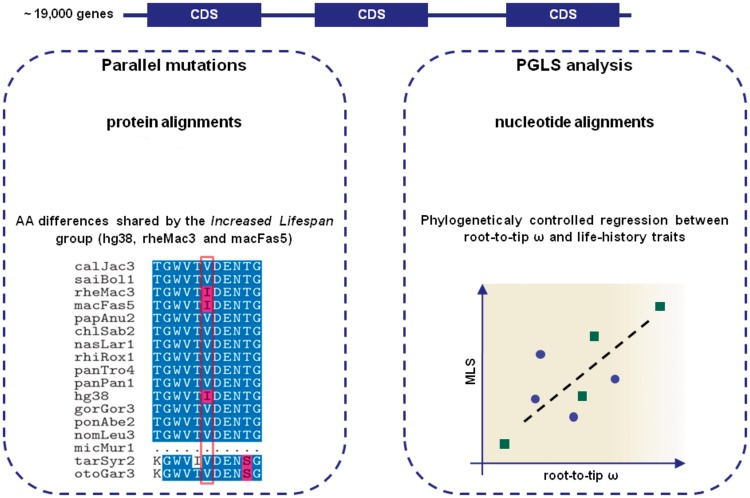
Fig. 1.
Illustrated workflow of the two tests used in this study for measuring variation associated to MLS. Parallel amino-acid (AA) changes were investigated across all coding-regions for presence in the Increased Lifespan group of primate species compared with the rest, as shown in the example with a shared “I” (left panel). The second approach (right panel), was planned to test for coevolution between rates of protein evolution and life-history traits (or in short, gene–phenotype coevolution). First, root-to-tip dN/dS (ω) ratios were calculated for each species, for each gene. Using a phylogenetically controlled method, we tested for a correlation between the log10-transformed rates of protein evolution (i.e., root-to-tip ω, x axis) and each life-history trait (e.g., MLS, y axis). A significant linear relationship between gene and phenotype provides support for coevolution, as hypothetically illustrated in the figure.