Fig. 1.
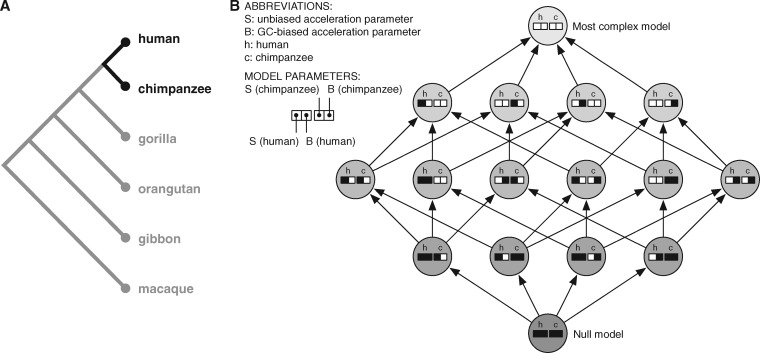
Testing for acceleration in five apes. To test a conserved noncoding element for an accelerated rate of unbiased and/or GC-biased substitutions in any of five apes, we perform a series of up to 1024 nested LRTs. (A) The apes tested and their phylogenetic relationships (branches not to scale). For illustrating the approach, consider the possible models for just the human and chimpanzee branches. (B) Example model selection procedure for two species. The null model (bottom node) has no unbiased acceleration (S = 0 on both human and chimpanzee branches) and no GC-biased acceleration (B = 0 on both branches). There are four models with one of the parameters not equal to zero (next set of four nodes): S > 0 in human (right-most), S > 0 in chimpanzee (second from right), B > 0 in human (second from left), and B > 0 in chimp (left-most). These are nested inside the six (i.e., 4 choose 2) possible models with two parameters constrained to zero, which are nested inside the four possible models with one parameter constrained to zero. The full model (top node) has all four parameters not constrained to zero. Our algorithm starts with the null model and performs the LRT corresponding to each subsequent arrow moving from the bottom to the top of the graph of possible models, stopping if none of the models with an additional parameter greater than zero has significantly higher likelihood than the most complex model visited. For all five primates, the graph of nested models has 1024 nodes and a similar structure.