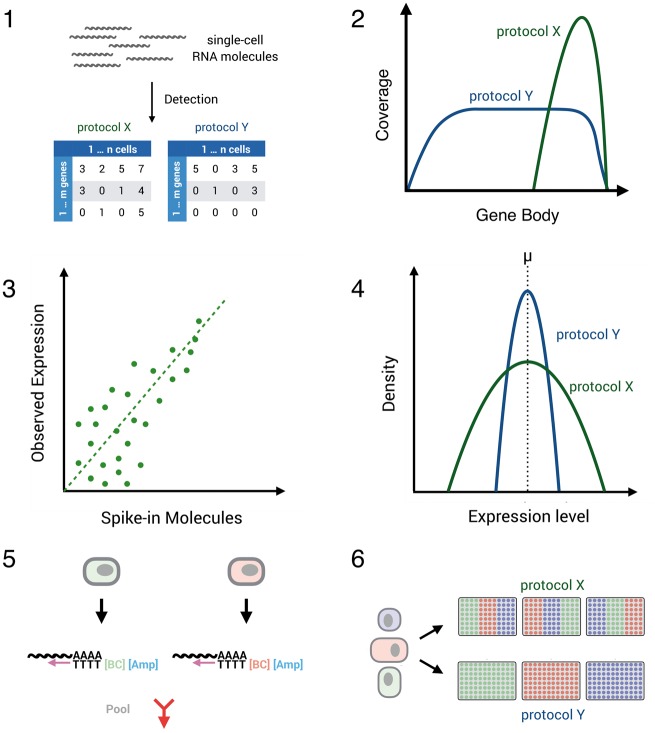
Figure 4.
Comparing scRNA-seq protocol performance. Several aspects determine the technical performance of a scRNA-seq protocol: (1) sensitivity of protocols to detect mRNA transcripts can be defined by the number of genes/transcripts (UMIs) per cell detected above stochastic noise. (2) Coverage of transcripts: with Smart-seq1/2, ideally the full length of the transcript is covered. Conversely, early barcoding and UMI-methods enrich for the 3′ and 5′ prime end of the sequences. (3) Accuracy of estimated expression levels reflect the correlation of known transcript concentrations and measured transcript expression. Notably, this correlation also depends on the sensitivity and precision of a method. (4) Precision of estimated expression levels reflects the measurement error of expression in single cells and depends on sensitivity and amplification noise. The latter is essentially abolished by UMIs. (5) The throughput of a method depends on the cell isolation method and on the costs per cell, which are strongly reduced by the depicted early barcoding (6) Batch effects of library generation can be a decisive factor for interpreting results, and methods that allow a balanced experimental design have a decisive advantage in this respect.