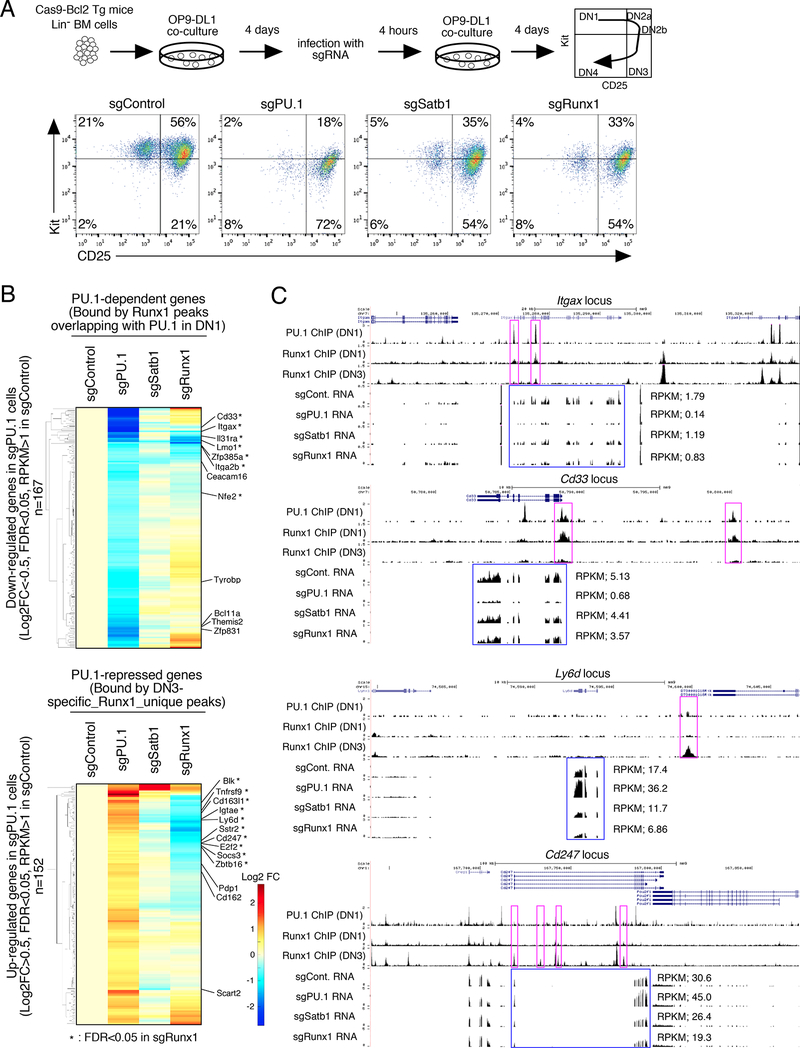
Figure 7. Satb1 and Runx1 roles in PU.1-mediated gene regulation in primary DN cells.
(A), Flow cytometric analysis of primary pro-T cells after CRISPR-Cas9-mediated knockout of PU.1, Satb1 or Runx1. (Upper) Experimental scheme. (Lower) sgRNA transduced BM-derived precursors after 8 days of OP9-DL1 culture. Representative of three independent experiments.
(B), Effects of Satb1 and Runx1 disruption on PU.1-mediated gene regulation in DN cells, by RNA-seq of transduced CD25+ DN cells from (A). Hierarchical clustering of expression changes in response to different treatments are shown, among genes down-regulated (top) and up- regulated (bottom) in PU.1-deficient DN cells (|Log2FC|>0.5, FDR<0.05, RPKM>1 in sgControl). Based on RNA-seq from independent duplicates.
(C), The binding patterns of PU.1 (DN1) and Runx1 (DN1 and DN3), and RNA-seq tracks are shown for Spi1 (PU.1), Satb1 and Runx1-deficient DN cells at the Itgax and Cd33 PU.1- dependent loci and at the Ly6d and Cd247 PU.1-repressed loci. Representative of two independent experiments. See also Fig. S6 and Table S5