Figure 2.
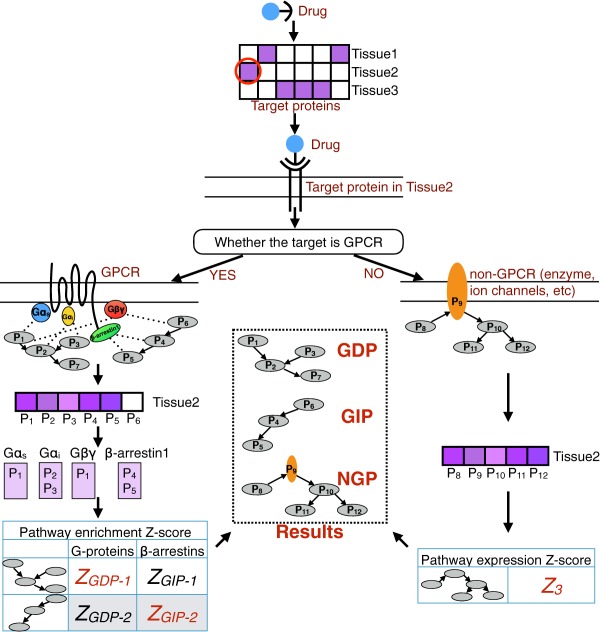
Workflow of Drugs to target pAthways by the Tissue Expression (DATE). A drug is first mapped to its target proteins using DrugBank. Then, tissue expression data are used to find the target protein that is highly expressed in each tissue. Next, two different processes were followed depending on whether the target protein is a G‐protein coupled receptor (GPCR) or not (GPCRs do not participate in the cellular activities directly, they pass the signals down to transducers). GPCRs are connected to downstream pathways using our previously developed method predicting GPCR downstream signaling pathways using the tissue expression (GOTE). In GOTE, the target GPCR is first mapped to the highly expressed transducers (G‐proteins or β‐arrestins) in the tissue. Then, for each transducer (G‐protein or β‐arrestin), a list of tissue‐specific binding proteins is obtained by combining BioGRID protein‐protein interaction data with the tissue expression of binding proteins. Pathway enrichment analysis is then performed based on the tissue‐specific binding proteins of each transducer using Fisher's exact test. For each pathway, the Z‐scores of all G‐proteins (or β‐arrestins) are combined into a single Z‐score using Stouffer's Method. Eventually, pathways with significant Z‐scores are connected to the drug in the tissue as G‐protein dependent pathways (GDPs; those that are associated with G‐proteins) or G‐protein independent pathways (GIPs; those that are associated with β‐arrestins). Non‐GPCRs are first connected to the annotated pathways. Then an expression Z‐score will be calculated for each annotated pathway to determine whether the pathway is highly expressed in the tissue, and the pathways with significant Z‐scores are connected to the drug in the tissue as non‐GPCR target pathways (NGPs).
