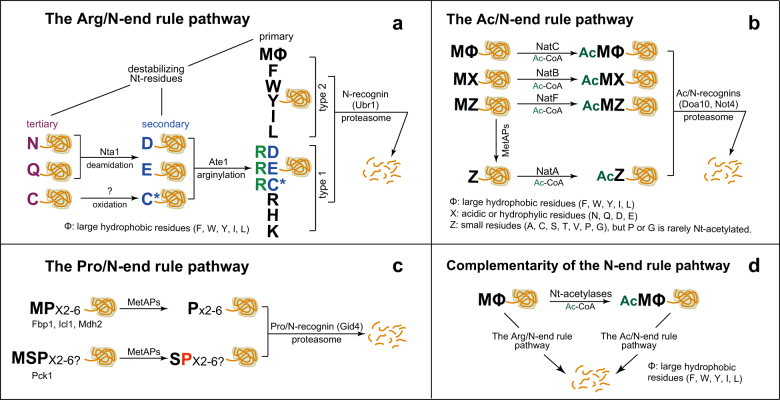
Fig. 1. The N-end rule pathway in S. cerevisiae.
a The conventional Arg/N-end rule pathway. Nta1 N-terminal amidase deamidates tertiary destabilizing N-terminal residues Asn and Gln into secondary destabilizing N-terminal residues Asp and Glu, respectively. Subsequently, Ate1 arginyl-transferase attaches Arg to the N-terminal Asp or Glu. Eventually, Ubr1 recognizes the type-1 or type-2 primary destabilizing N-terminal residues of the target substrates for polyubiquitylation and subsequent proteasomal degradation. Note that the N-terminal Cys oxidation-dependent degradation pathway has not yet been established in yeast. b The Ac/N-end rule pathway. Met aminopeptidases (MetAPs) remove the N-terminal initiator Met (iMet), if the residues smaller than Val occupy the penultimate position. NatA, NatB, or NatC Nt-acetylates target proteins largely based on their first two residues. The Ac/N-recognins (Doa10 and Not4) specifically recognize the N-terminal acetyl moiety of the N-terminally acetylated proteins for polyubiquitin-mediated and proteasome-dependent degradation. c The Pro/N-end rule pathway. N-terminal Pro or N-terminal Ser with Pro at position 2 can work as Pro/N-degrons for Pro/N-recognin Gid4 in the GID Ub ligase complex. d Functional and mechanical interplay between the two branches of the N-end rule pathway