Abstract
Highly-lignified culms of bamboo show distinctive anatomical and mechanical properties compared with the culms of other grass species. A cell culture system for Phyllostachys nigra has enabled investigating the alterations in cellular states associated with secondary cell wall formation during its proliferation and lignification in woody bamboos. To reveal transcriptional changes related to lignification in bamboo, we analyzed transcriptome in P. nigra cells treated with the synthetic auxin 2,4-dichlorophenoxyacetic acid (2,4-D) and the synthetic cytokinin benzylaminopurine (BA) by RNA-seq analysis. We found that some genes putatively involved in cell wall biogenesis and cell division were up-regulated in response to the 2,4-D treatment, and the induction of lignification by the BA treatment was correlated with up-regulation of genes involved in the shikimate pathway. We also found that genes encoding MYB transcription factors (TFs) show correlated expression patterns with those encoding cinnamyl alcohol dehydrogenase (CAD), suggesting that MYB TFs presumably regulate secondary cell wall formation in the bamboo cells. These findings suggest that cytokinin signaling may regulate lignification in P. nigra cells through coordinated transcriptional regulation and metabolic alterations. Our results have also produced a useful resource for better understanding of secondary cell wall formation in bamboo plants.
Introduction
Bamboo is an ecologically and economically important grass species. It belongs to the largest subfamily, the Bambusoideae, in the grass family (Poaceae)1,2, which contains more than 1,500 species that are adapted to diverse climates. It has been exploited for a range of uses such as food, medicine, charcoal, and housing materials, especially in Asia3. Owing to their wide utility and productivity, bamboo species are increasingly regarded as a valuable resource for use in renewable energy in the development of a low-carbon society4,5.
It is well known that bamboo presents unique biological properties in its vegetative growth and sexual reproduction. It has a rhizome system for lateral growth and forms highly lignified woody culms for longitudinal growth without secondary growth, which are its distinguishing characteristics compared with other grass species and tree species. Moreover, bamboo species often have flowering intervals from several to more than a hundred years, which is another characteristic feature of the sexual reproduction of bamboo species. To elucidate gene regulatory networks involved in these biological phenomena observed in bamboo species, several studies have utilized transcriptome analyses, and identified spatiotemporal expressions of genes explored across different tissues and developmental stages6–9, which improved the understanding of the molecular mechanisms underlying the development and growth in bamboo. However, these analyses provided little information at the cellular level, and did not identify the molecular mechanisms of cellular differentiation associated with its highly-lignified culm formation.
Cell culture systems have been established in some model plant species, such as Arabidopsis T8710 and tobacco BY-211, and exploited to investigate a wide range of aspects of plant cell biology. Recently, Ogita et al. established a novel xylogenic suspension culture approach in the bamboo Phyllostachys nigra (resource number in RIKEN BioResource Center; rpc00047) that enabled investigation of lignification in living bamboo cells12. The cultured P. nigra cells showed cell wall thickening and proliferation in response to treatment with the synthetic auxin 2,4-dichlorophenoxyacetic acid (2,4-D), and lignification occurred in response to treatment with the synthetic cytokinin benzylaminopurine (BA). After 3–5 days of induced lignification, the cells showed xylogenic differentiation, the presence of fiber-like elements with cell wall thickening, and tracheary elements with formation of perforations12. Elucidation of the global gene expression profiles of the suspension culture cells under lignification conditions should allow identification of the gene groups important to this process and enable the characterization of gene networks involved in lignification.
The highly conserved genic regions among Phyllostachys species suggest that the draft genome sequence of P. edulis (moso bamboo)13 can provide a reference genome sequence for RNA-seq-based transcriptome analyses to investigate gene expression patterns in related bamboo species whose whole genome sequences have not yet been deciphered14. In-depth analysis of the transcriptome dynamics in response to induced lignification in bamboo cells will provide new insights into the molecular basis of cellular differentiation.
In this study, we aimed to reveal the transcriptional regulatory networks underlying the lignification process of bamboo at the cellular level. We used RNA-seq based transcriptome analysis to obtain an overview of the gene expression of cultured P. nigra cells, rpc00047, and sought to identify the key pathways and transcription factors involved in its lignification process.
Results and Discussion
Overview of the transcriptome analysis of P. nigra cells
We sequenced mRNAs from control and treated P. nigra cells, and found that almost all of the filtered reads could be mapped to the P. edulis draft genome. The P. nigra cells were cultured with treatments of either 2,4-D or BA, and sampled at four and seven days after the initiation of the treatments. Although the cross-platform assessments suggested that Illumina and Ion Torrent would present approximately similar results in RNA-seq based transcriptome profiling, each of them could have platform-specific differentially expressed genes15. To minimize biases between the platforms, we applied the Illumina and Ion Torrent sequencing platforms for our RNA-seq analysis of P. nigra cells. From the sequenced mRNAs, we obtained 783 million reads amounting to approximately 78 gigabases in the filtered dataset; 93.22% of these sequences mapped to the P. edulis draft genome (Supplementary Table S1). Thus, even though we used the P. edulis draft genome13 as the reference sequence, we obtained a high rate of successful mapped reads suggesting that the P. edulis draft genome provides a useful reference genome sequence to analyze transcriptomes in bamboo species, probably due to their conserved genic sequences. We identified 25,443 P. nigra genes significantly expressed in the cells (at least one condition with average RPM values of replicated samples ≥1), which are corresponding to the counterparts annotated in the P. edulis draft genome. These results indicate that, in the P. nigra cells, genes corresponding to as much as 80% of the genes annotated in the P. edulis genome are detectable as significantly expressed genes (Supplementary Fig. 1a). Comparison of datasets from two duplicate samples after seven days BA treatment and sequenced on the Illumina platform gave Pearson’s correlation coefficients (PCC) of up to 0.996. Additionally, comparison of datasets from the same sample conditions using the two sequencing platforms gave high PCC values (e.g., 0.930 between control conditions); the slightly lower PCC values across sequence platforms likely reflect differences in the sequencing methodologies (Supplementary Fig. S1b). To our knowledge, this is the first study of deep transcriptome analyses of P. nigra, and the data from the study serve as a resource of P. nigra transcripts, which offer clues to identifying genes related to cellular differentiation and lignification in bamboo.
Expression of monolignol pathway genes in response to hormonal treatment of P. nigra cells
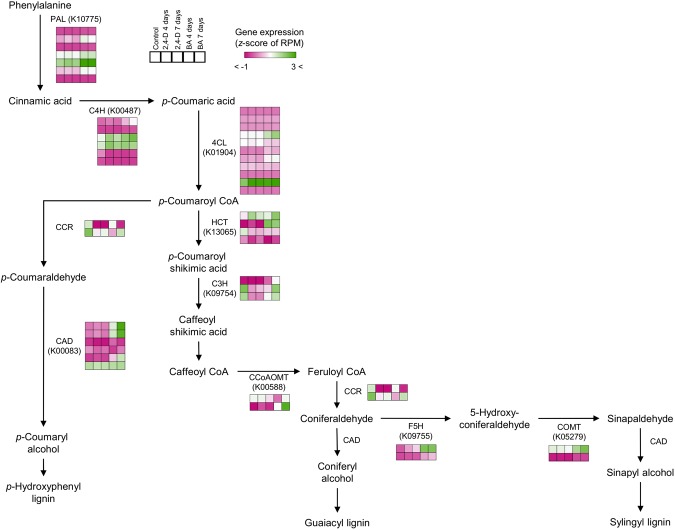
An expression analysis of monolignol pathway genes in P. nigra showed expression of genes putatively involved in the lignification process in the cultured cells. In our previous observation, the P. nigra cells treated with auxins such as 2,4-D or picloram showed increased cell division and suppression of lignification, whereas cells treated with BA showed induced lignification12. Moreover, the P. nigra cells under the BA treatment presented increased signals of phloroglucinol-HCl, indicating induction of lignification, and found transcriptional changes in some xylogenesis-related genes including PAL, C4H, CCoAOMT, and CCR induced at day 4 of treatment with BA12. To reveal the transcriptional differences underlying the cellular responses against these hormonal treatments observed in the P. nigra cells, we assessed the expression patterns of P. nigra genes putatively involved in monolignol biosynthesis. We found that P. nigra genes encoding CCR and C3H were down-regulated in response to 2,4-D treatment, and that some downstream genes in the monolignol biosynthesis pathway, such as CAD, F5H, and COMT, were up-regulated in response to BA treatment (Fig. 1). Specifically, we found that three genes putatively encoding CAD (homologous to PH01000043G2130, PH01000043G2150, and PH01003504G0010 in P. edulis), F5H (homologous to PH01000012G2270 in P. edulis), and COMT (homologous to PH01000383G0390 in P. edulis) showed a clear response to BA treatment, suggesting their coordinated gene expressions associated with cellular lignification in P. nigra cells. We also found that some genes, such as those encoding PAL, C4H, and 4CL, were up-regulated in response to both 2,4-D and BA treatments. These results suggest that the specific up-regulation of genes encoding CAD, F5H, and COMT in response to BA treatment may presumably be molecular differences associated with the differential cellular responses. For some copies in each gene group, our results are consistent with the cellular responses that initiate differentiation and lignification as well as the expression patterns of genes investigated in the previous study of P. nigra cells12. We also found some gene copies, even those encoding the same enzyme that showed different patterns of expression and/or a low level of expression in all conditions, suggesting that subfunctionalization and/or nonfunctionalization may have caused diversification of the expression patterns of these putative paralogous genes. Through our transcriptome analysis, we identified genes involved in the monolignol pathway in P. nigra that were expressed consistently with lignification, suggesting these genes and orthologs will be useful expression markers for monitoring the lignification process in bamboo species.
Figure 1.
Expression of P. nigra genes involved in the monolignol pathway. P. edulis genes encoding phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarate-CoA ligase (4CL), hydroxycinnamoyl CoA:shikimate transferase (HCT), p-coumarate 3-hydroxylase (C3H), caffeoyl CoA O-methyltransferase (CCoAOMT), cinnamoyl CoA reductase (CRR), ferulate 5-hydroxylase (F5H), caffeic acid O-methyltransferase (COMT), and cinnamyl alcohol dehydrogenase (CAD) were represented in the monolignol pathway. The expression patterns of P. nigra genes corresponding to their homologs in P. edulis were estimated from the RPM values obtained from the cross-species mapping of P. nigra RNA-seq reads to the P. edulis genome. The color gradient represents normalized gene expression based on z-score of the RPM values.
P. nigra gene expression in response to 2,4-D and BA treatments
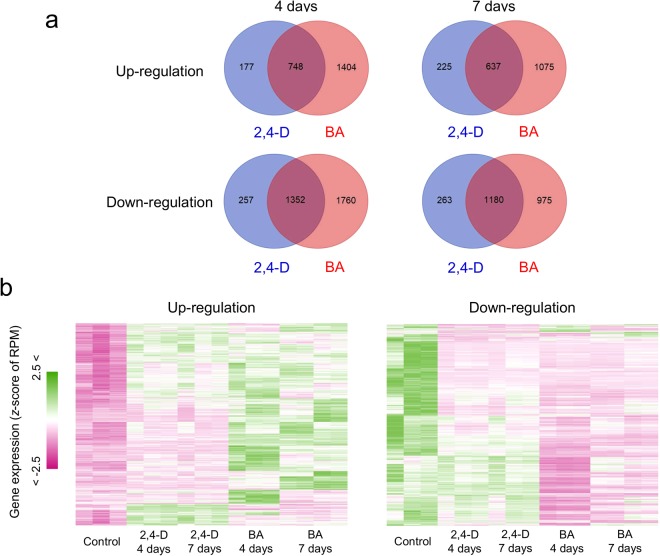
Comparing the gene expression in cells cultured under control and hormone-treated conditions, we identified a number of DEGs in the latter treatments. We first sought to identify the genes whose expression was responsive to 2,4-D and BA treatments by comparing up- and down-regulated genes. Our results showed that BA treatment triggered a change in expression of a larger number of genes than the 2,4-D treatment (Fig. 2a). Using the threshold adjusted p-value < 1e-03, 1,404 genes were found to be specifically up-regulated in samples from the 4-day treatment with BA compared with that in the control, whereas 177 genes were specifically up-regulated in 4-day treatments with 2,4-D; in addition, 748 genes were up-regulated in both treatment groups (Fig. 2a). In view of clustered gene expression patterns among DEGs, we found that more genes respond significantly to BA than to 2,4-D (Fig. 2b). These results indicate that many genes show altered expression patterns in response to the hormone treatments in P. nigra cells, which suggests that a broad range of cellular systems are influenced by the hormone treatments.
Figure 2.
Differentially expressed genes (DEGs) in P. nigra cells in response to the 2,4-D and BA treatments. (a) Venn diagrams representing the number of up-regulated and down-regulated DEGs in each of the hormonal conditions. (b) Heat maps of a hierarchical clustering analysis based on the average linkage method of gene expression profiles across samples using the up-regulated and down-regulated genes. The color gradient represents normalized gene expression based on the z-score of the RPM values.
Functional classification of the DEGs
Through enrichment analysis of functional classes and pathways among the DEGs up-regulated in response to the 2,4-D and BA treatments, we assessed the cellular functions that might be associated with the cellular differentiation induced by the hormone treatments in the P. nigra cultured cells. Among the DEGs specifically up-regulated in response to the 2,4-D treatment, we found that the genes encoding galactose transferase (MapMan #10.3.1.1) and cellulose synthase (MapMan #10.2.1) were enriched in both the 4-day and 7-day treatments (Table 1). We also found that genes putatively encoding cellulose synthase A (KEGG: K10999) were enriched with the 7-day treatment. These findings indicate that P. nigra cells treated with 2,4-D induce expression of the genes related to cell wall biogenesis, which is consistent with our previous observation of thickening and proliferation of the cells in response to the 2,4-D treatment. We also found that laccase activity (KEGG: K05909) was an enriched function among DEGs in response to the 2,4-D treatment. Because secondary wall-associated laccases are required for lignification by catalyzing oxidation of phenolic compounds16,17, the 2,4-D treatments may partially activate the process for secondary cell wall formation in P. nigra cells. Among the DEGs specifically up-regulated in response to the BA treatment, we found some enriched functions related to genes encoding enzymes involved in amino acid biosynthesis. Specifically, we found an enrichment of genes involved in the shikimate pathway18 for biosynthesis of aromatic amino acids (MapMan: #13.1.6.5.1, #13.1.6.5.5, and #13.1.6.1.1, KEGG: K01626), suggesting specific activation of the shikimate pathway in response to the BA treatment (Fig. 3), which can occur prior to monolignol biosynthesis (Fig. 1) and the subsequent lignification observed in the P. nigra cells. We also found over-representation of the genes related to transporter activities in the DEGs upregulated in response to the BA treatment (MapMan: #34.99, #34.15, and #34.16, KEGG: K03301 and K03549) (Table 2), suggesting that BA treatment activates genes encoding transporters and subsequently affects cellular logistics in the P. nigra cells. The list of upregulated DEGs classified to the MapMan binode with the prefix #34 (transport) has showcased genes homologous to various types of transporters, including 12 genes homologous to ATP-binding cassette (ABC) transporters (Supplementary Table S2), which may be involved in the transportation of monolignols19. Specifically, four genes encoding putative G family ABC transporters (homologous to PH01000231G0750, PH01002712G0070, PH01002800G0200, and PH01003385G0160 in P. edulis) might be involved in transporting monolignols from the cytoplasm to the cell wall for polymerization in the P. nigra cells. In Arabidopsis, a member of G family ABC transporter, AtABCG29, shows p-coumaryl alcohol transporter activity, and is the first monolignol transporter reported20,21. More recently, expression analysis of transporter encoding genes during tracheary element differentiation in cultured Arabidopsis cells suggested that four Arabidopsis ABC transporters; AtABCG11, AtABCG22, AtABCG36, and AtABCG29, may also be involved in lignification as candidate monolignol transporters22. The P. nigra cell culture system will provide a useful resource to identify ABC transporters that regulate cellular localization of monolignols in bamboo species, which may offer us novel insights into the evolution of the monolignol biosynthetic pathway in higher plants. In the DEGs upregulated in response to the BA treatment, we also found significant enrichment of a number of genes classified into an unknown functional category (MapMan: #35.2) (Table 2), suggesting that the BA treatment may affect the expression of genes involved in various cellular functions that remain unexplored. On the whole, these results illuminate the transcriptional alterations of P. nigra cells in response to both the 2,4-D and BA treatments, providing a comprehensive list of genes that may be involved in cellular functions related to proliferation and lignification (Supplementary Tables S3 and S4).
Table 1.
Enriched functions found in the up-regulated genes under the 2,4-D condition in the P. nigra cells.
| Days | Ontology | Description | P-value | Resources |
|---|---|---|---|---|
| 4 days | 10.3.1.1 | cell wall.hemicellulose synthesis.xyloglucan.XXXG galactose Transferase | 9.10E-05 | MapMan |
| 27.3.63 | RNA.regulation of transcription.PHD finger transcription factor | 0.000107 | ||
| 30.11 | signalling.light | 0.001734 | ||
| 10.2.1 | cell wall.cellulose synthesis.cellulose synthase | 0.002123 | ||
| 35.2 | not assigned.unknown | 0.002507 | ||
| K11665 | DNA helicase INO80 [EC:3.6.4.12] | 3.05E-05 | KEGG | |
| K12619 | 5′-3′ exoribonuclease 2 [EC:3.1.13.-] | 0.000181 | ||
| 7 days | 10.2.1 | cell wall.cellulose synthesis.cellulose synthase | 8.39E-07 | MapMan |
| 27.3.63 | RNA.regulation of transcription.PHD finger transcription factor | 5.28E-06 | ||
| 10.3.1.1 | cell wall.hemicellulose synthesis.xyloglucan.XXXG galactose Transferase | 0.000147 | ||
| 11.1.1.1 | lipid metabolism.FA synthesis and FA elongation.Acetyl CoA Carboxylation.homomeric Enzyme | 0.000293 | ||
| 35.1.12 | not assigned.no ontology.pumilio/Puf RNA-binding domain-containing protein | 0.000548 | ||
| 13.1.3.1.1 | amino acid metabolism.synthesis.aspartate family.asparagine.asparagine synthetase | 0.000725 | ||
| 31.1.1.3.8 | cell.organisation.cytoskeleton.Myosin.Class VII | 0.000725 | ||
| 35.2 | not assigned.unknown | 0.000897 | ||
| 11.9.3.3 | lipid metabolism.lipid degradation.lysophospholipases.glycerophosphodiester phosphodiesterase | 0.00424 | ||
| 29.2.2.3.1 | protein.synthesis.ribosome biogenesis.Pre-rRNA processing and modifications.snoRNPs | 0.00424 | ||
| K18442 | brefeldin A-inhibited guanine nucleotide-exchange protein | 1.87E-05 | KEGG | |
| K10999 | cellulose synthase A [EC:2.4.1.12] | 2.27E-05 | ||
| K11665 | DNA helicase INO80 | 4.93E-05 | ||
| K13462 | guanine nucleotide-exchange factor | 4.93E-05 | ||
| K17943 | pumilio RNA-binding family | 0.000147 | ||
| K12879 | THO complex subunit 2 | 0.000147 | ||
| K11262 | acetyl-CoA carboxylase/biotin carboxylase 1 [EC:6.4.1.2, EC:6.3.4.14, EC:2.1.3.15] | 0.000293 | ||
| K01953 | asparagine synthase (glutamine-hydrolysing) [EC:6.3.5.4] | 0.000486 | ||
| K01090 | protein phosphatase [EC:3.1.3.16] | 0.001341 | ||
| K12617 | DNA topoisomerase 2-associated protein PAT1 | 0.001341 | ||
| K05909 | laccase [EC:1.10.3.2] | 0.003104 |
Figure 3.
Expression of P. nigra genes involved in shikimate acid and phenylalanine biosynthesis. The P. edulis genes encoding 2-dehydro-3-deoxyphosphoheptonate aldolase (DAHPS), dehydroquinate synthase (DHQS), DHQD/SD (3-dehydroquinate dehydratase/shikimate-NADP oxidoreductase), shikimate kinase (SK), 5-enolpyruvylshikimate-3-phosphate synthase (ESPS), chorismate synthase (CS), chorismate mutase (CM), and arogenate dehydratase (ADT) were represented in the shikimate acid and phenylalanine pathways. KEGG Orthology IDs are also shown. The color gradient represents normalized gene expression based on the z-score of the RPM values.
Table 2.
Enriched functions found in the up-regulated genes under the BA condition in the P. nigra cells.
| Days | Ontology | Description | P-value | Resources |
|---|---|---|---|---|
| 4 days | 35.2 | not assigned.unknown | 2.20E-16 | MapMan |
| 34.99 | transport.misc | 2.11E-05 | ||
| 30.2.6 | signalling.receptor kinases.leucine rich repeat VI | 9.75E-05 | ||
| 8.1.5 | TCA/org transformation.TCA.2-oxoglutarate dehydrogenase | 0.000116 | ||
| 13.1.6.5.1 | amino acid metabolism.synthesis.aromatic aa.tryptophan.anthranilate synthase | 0.000224 | ||
| 13.1.3.4.11 | amino acid metabolism.synthesis.aspartate family.methionine.S-adenosylmethionine synthetase | 0.000327 | ||
| 35.1.1 | not assigned.no ontology.ABC1 family protein | 0.000336 | ||
| 29.2.4 | protein.synthesis.elongation | 0.000339 | ||
| 13.1.6.5.5 | amino acid metabolism.synthesis.aromatic aa.tryptophan.tryptophan synthase | 0.00039 | ||
| K03301 | ATP:ADP antiporter, AAA family | 5.16E-05 | KEGG | |
| K03327 | multidrug resistance protein, MATE family | 5.53E-05 | ||
| K00799 | glutathione S-transferase [EC:2.5.1.18] | 0.000149 | ||
| K00600 | glycine hydroxymethyltransferase [EC:2.1.2.1] | 0.000224 | ||
| K04043 | molecular chaperone DnaK | 0.000224 | ||
| K13024 | inositol-hexakisphosphate/diphosphoinositol-pentakisphosphate 1-kinase [EC:2.7.4.24] | 0.000224 | ||
| K00164 | 2-oxoglutarate dehydrogenase E1 component [EC:1.2.4.2] | 0.000327 | ||
| K07513 | acetyl-CoA acyltransferase 1 [EC:2.3.1.16] | 0.000327 | ||
| K10592 | E3 ubiquitin-protein ligase HUWE1 [EC:2.3.2.26] | 0.000327 | ||
| K13034 | L-3-cyanoalanine synthase/cysteine synthase [EC:2.5.1.47, EC:4.4.1.9] | 0.000327 | ||
| K14492 | two-component response regulator ARR-A family | 0.00039 | ||
| 7 days | 35.2 | not assigned.unknown | 2.20E-16 | MapMan |
| 12.2.1.1 | N-metabolism.ammonia metabolism.glutamate synthase.ferredoxin dependent | 1.27E-06 | ||
| 13.1.6.1.1 | amino acid metabolism.synthesis.aromatic aa.chorismate.3-deoxy-D-arabino-heptulosonate 7-phosphate synthase | 6.17E-06 | ||
| 34.99 | transport.misc | 8.99E-05 | ||
| 13.1.3.4.11 | amino acid metabolism.synthesis.aspartate family.methionine.S-adenosylmethionine synthetase | 0.000148 | ||
| 30.2.6 | signalling.receptor kinases.leucine rich repeat VI | 0.000184 | ||
| 34.15 | transport.potassium | 0.000202 | ||
| 25.1 | C1-metabolism.glycine hydroxymethyltransferase | 0.000227 | ||
| 34.16 | transport.ABC transporters and multidrug resistance systems | 0.000319 | ||
| K00284 | glutamate synthase (ferredoxin) [EC:1.4.7.1] | 1.27E-06 | KEGG | |
| K01626 | 3-deoxy-7-phosphoheptulonate synthase [EC:2.5.1.54] | 6.17E-06 | ||
| K03327 | multidrug resistance protein, MATE family | 8.13E-06 | ||
| K01278 | dipeptidyl-peptidase 4 [EC:3.4.14.5] | 3.79E-05 | ||
| K03549 | KUP system potassium uptake protein | 3.88E-05 | ||
| K00600 | glycine hydroxymethyltransferase [EC:2.1.2.1] | 7.97E-05 | ||
| K13034 | L-3-cyanoalanine synthase/cysteine synthase [EC:2.5.1.47, EC:4.4.1.9] | 0.000148 | ||
| K01904 | 4-coumarate–CoA ligase [EC:6.2.1.12] | 0.000347 | ||
| K00789 | S-adenosylmethionine synthetase [EC:2.5.1.6] | 0.00036 | ||
| K01783 | ribulose-phosphate 3-epimerase [EC:5.1.3.1] | 0.00036 |
Changes in expression of transcription factor genes in response to BA treatment in P. nigra cells
We identified transcription factors (TFs) possibly involved in cellular lignification by a comparison of the expression patterns of P. nigra genes putatively encoding transcription factors with those of genes encoding enzymes that catalyze downstream processes in the monolignol pathways, such as CAD, F5H, and COM. This comparison yielded 1,663 genes that putatively encode DNA-binding domains (DBDs) in genes from 60 TF families that are annotated in the P. edulis genome. In addition, based on a comparison of P. edulis and Arabidopsis genomes, we identified genes homologous to Arabidopsis TFs for the promoters involved in cellulose, xylan, and lignin biosynthesis during secondary cell wall formation. These Arabidopsis genes were identified by a yeast one-hybrid assay23. Based on the co-expression patterns of these TFs with genes encoding enzymes involved in downstream monolignol pathways, we identified 18 genes putatively encoding TFs, including 7 MYB family genes, three ERF/AP2 family genes, two calmodulin-binding transcription activator (CAMTA), two GRAS family genes, and one bHLH family gene, which showed co-expression patterns with genes putatively encoding CAD, F5H or COMT (PCC ≥ 0.8) (Table 3). We found that the TF genes were homologous to the AtMYB85 and AtMYB20 genes of Arabidopsis. AtMYB85 is a known lignin-specific transcription factor that regulates lignin biosynthesis genes to activate secondary cell wall formation in Arabidopsis24,25. AtMYB20 also regulates secondary cell wall biosynthesis and is induced by NAC transcription factors that regulate secondary cell wall biosynthesis such as SND1, NSTs, and VNDs in Arabidopsis24. A co-expression network analysis of genes expressed during internode development in rice identified orthologs of MYB85 and MYB20 as important for secondary cell wall development26. These findings suggest that the transcriptional regulatory network for secondary cell wall formation in P. nigra cells might include some TFs conserved between dicot and monocot plants. Moreover, genes in the ERF/AP2 family are homologous to RAP2.12 in Arabidopsis, which is known to have a role in ethylene signaling27,28, and possibly regulates the final stages of xylogenesis through ethylene signaling29,30. The possible activation of genes for xylogenesis after the induction of lignification in bamboo cells suggests that secondary cell wall formation and subsequent xylogenesis might be coordinated through CK/ethylene crosstalk in bamboo cells31,32. We also identified one gene encoding bHLH transcription factors that showed correlated expression with genes for monolignol biosynthesis. This gene was homologous to bHLH105, which encodes IAA-LEUCINE RESISTANT3 (ILR3) that has a crucial role in Fe homeostasis through direct interaction with bHLH34 and bHLH10433. It has been reported that both bHLH transcription factors participate in an Arabidopsis gene regulatory network for secondary cell wall biosynthesis23,34.
Table 3.
Transcription factors whose gene expression patterns correlated with the genes involved in monolignol biosynthesis in P. nigra.
| IDs of moso bamboo homologs | Closest homologs | Promoters | Gene expression (z-score of RPM) | Correlation coefficients with expression patterns of P. nigra genes involved in the monolignol biosynthesis | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rice | Arabidopsis | Gene symbols in Arabidopsis | cellulose | lignin | xylan | Control | 2,4-D 4 days | 2,4-D 7 days | BA 4 days | BA 7 days | CAD | CAD | CAD | F5H | COMT | |
| PH01000030G0050 | LOC_Os04g50770 | AT1G79180 | MYB63 | ✔ | ✔ | 0.39 | −1.17 | −1.21 | 0.89 | 1.10 | 0.68 | 0.72 | 0.68 | 0.41 | 0.83 | |
| PH01000060G0800 | LOC_Os09g36250 | AT4G22680 | MYB85 | ✔ | ✔ | −0.61 | −0.59 | −0.54 | −0.23 | 1.98 | 0.94 | 0.90 | 0.92 | 0.69 | 0.87 | |
| PH01000150G0510 | LOC_Os08g39830 | AT1G73730 | EIL3 | ✔ | −1.69 | −0.35 | 0.36 | 1.32 | 0.36 | 0.56 | 0.56 | 0.58 | 0.90 | 0.42 | ||
| PH01000210G1070 | LOC_Os02g54160 | AT1G53910 | RAP2.12 | ✔ | −0.69 | −0.89 | −0.65 | 0.48 | 1.75 | 0.99 | 0.98 | 0.98 | 0.83 | 0.96 | ||
| PH01000348G0830 | LOC_Os05g38140 | AT5G54680 | bHLH105 | ✔ | ✔ | ✔ | −0.13 | −1.10 | −0.94 | 0.58 | 1.60 | 0.91 | 0.92 | 0.90 | 0.66 | 0.96 |
| PH01000847G0490 | LOC_Os09g23620 | AT1G66230 | MYB20 | ✔ | −0.84 | −0.77 | −0.83 | 1.16 | 1.29 | 0.94 | 0.96 | 0.95 | 0.91 | 0.95 | ||
| PH01001102G0050 | LOC_Os06g09390 | AT1G53910 | RAP2.12 | ✔ | −1.22 | −0.25 | −0.35 | 0.00 | 1.83 | 0.91 | 0.87 | 0.90 | 0.85 | 0.77 | ||
| PH01001197G0410 | LOC_Os10g22430 | AT5G48150 | PAT1 | ✔ | −1.38 | −0.17 | −0.53 | 1.60 | 0.47 | 0.70 | 0.73 | 0.73 | 0.92 | 0.66 | ||
| PH01001287G0090 | LOC_Os04g43680 | AT1G06180 | MYB13 | ✔ | −1.10 | −0.12 | −0.90 | 1.66 | 0.47 | 0.73 | 0.78 | 0.77 | 0.87 | 0.75 | ||
| PH01001342G0270 | LOC_Os06g14670 | AT1G66230 | MYB20 | ✔ | −0.92 | −0.91 | −0.48 | 1.62 | 0.69 | 0.81 | 0.84 | 0.82 | 0.92 | 0.80 | ||
| PH01001360G0240 | LOC_Os03g09100 | AT5G64220 | CAMTA2 | ✔ | −1.31 | −0.43 | −0.10 | 0.08 | 1.76 | 0.90 | 0.86 | 0.89 | 0.87 | 0.74 | ||
| PH01001360G0260 | LOC_Os03g09100 | AT5G64220 | CAMTA2 | ✔ | −1.61 | −0.11 | 0.22 | −0.04 | 1.54 | 0.76 | 0.72 | 0.76 | 0.82 | 0.57 | ||
| PH01001873G0040 | LOC_Os12g39220 | AT1G24625 | ZFP7 | ✔ | ✔ | −0.84 | −0.68 | −0.44 | 1.91 | 0.05 | 0.61 | 0.66 | 0.64 | 0.84 | 0.63 | |
| PH01002680G0080 | LOC_Os06g14670 | AT1G66230 | MYB20 | ✔ | −0.73 | −1.09 | −0.59 | 1.35 | 1.05 | 0.87 | 0.89 | 0.87 | 0.87 | 0.88 | ||
| PH01003093G0130 | LOC_Os09g36250 | AT4G22680 | MYB85 | ✔ | ✔ | −0.47 | −0.72 | −0.84 | 0.15 | 1.88 | 0.97 | 0.96 | 0.96 | 0.71 | 0.97 | |
| PH01003592G0180 | LOC_Os03g47140 | AT2G22840 | GRF1 | ✔ | ✔ | −1.75 | 0.71 | −0.47 | 0.59 | 0.92 | 0.64 | 0.64 | 0.67 | 0.81 | 0.52 | |
| PH01003923G0110 | LOC_Os01g12440 | AT1G50640 | ERF3 | ✔ | ✔ | −1.35 | −0.58 | −0.29 | 1.49 | 0.73 | 0.78 | 0.79 | 0.80 | 0.98 | 0.70 | |
| PH01004866G0030 | LOC_Os10g22430 | AT5G48150 | PAT1 | ✔ | −1.86 | 0.20 | −0.06 | 0.89 | 0.83 | 0.67 | 0.67 | 0.70 | 0.91 | 0.53 | ||
Metabolic differences of P. nigra cells treated with 2,4-D and BA
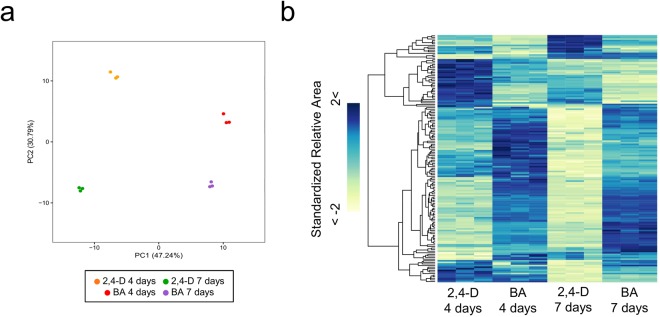
In the xylogenic suspension culture, P. nigra cells present differential metabolomic properties during proliferation and lignification in response to treatment with 2,4-D and BA. To reveal metabolic changes and explore its relationship with the transcriptional changes occurring during cellular differentiation, we performed a widely targeted metabolome analysis using CE-MS, with samples of P. nigra cells from the 4-day and 7-day treatments with 2,4-D and BA, respectively, and obtained a metabolome profile dataset composed of accumulation patterns of 214 compounds (Supplementary Table S5). In the widely targeted metabolome dataset, we found that amino acids synthesized through the shikimate pathway, such as phenylalanine (C_0075: Phe in Supplementary Table S6), tryptophan (C_0102: Trp in Supplementary Table S6), and tyrosine (C_0088: Tyr in Supplementary Table S6), are significantly increased their relative size of MS peaks under the BA conditions (p-value of Welch’s t-test < 0.001), suggesting that BA activates the shikimate pathway (Fig. 3), and consequently increases phenylalanine and tyrosine for monolignol biosynthesis in the P. nigra cells (Fig. 2). Moreover, we observed clear metabolic differences between the 4 sample conditions (Fig. 4a,b), suggesting significant metabolic alteration during the cellular differentiation process. Comparing the metabolome profiles of the P. nigra cells treated with 2,4-D and BA, we identified 120 and 131 metabolites that were differentially accumulated in the cells from the 4-day and 7-day treatments (p-value of Welch’s t-test <0.05), respectively, and found that metabolites of amino acids, nucleotide, sugars, and lipids were abundantly accumulated in the cells treated with BA (Supplementary Table S6). Based on the findings of our transcriptome analysis as well as widely-targeted metabolome analysis of the P. nigra cells treated with 2,4-D and BA, we obtained a comprehensive view of the transcriptomic and metabolic alterations occurring in response to the hormonal treatments, which induce proliferation and lignification in a bamboo species (Fig. 5). In response to the 2,4-D treatment, the P. nigra cells activate genes related to cell division and cell growth to promote their proliferation. During this process, they also activate genes associated with biosynthesis of cellulose and hemicellulose, which promote cell wall thickening through primary cell wall formation. In contrast, with BA treatment, P. nigra cells activate genes encoding TFs associated with secondary cell wall formation and the shikimate pathway to synthesize aromatic amino acids, followed by monolignol pathway genes to synthesize monolignol precursors.
Figure 4.
Metabolomic differences in P. nigra cells treated with 2,4-D and BA. (a) PCA of the metabolomic data of the P. nigra cells from the 4-day and 7-day treatments with 2,4-D and BA. (b) Hierarchically clustered heat map representation of 218 metabolites (lines) across the 4 conditions.
Figure 5.
Summary of cellular and transcriptomic alterations in P. nigra cells in response to the 2,4-D and BA treatments.
Conclusions
Our transcriptome analysis of cultured P. nigra cells that had been induced to undergo proliferation and lignification, identified changes to transcriptional regulatory networks and cellular metabolism, which were presumably related. Functional analyses of the genes encoding TFs that might be involved in lignification in P. nigra will undoubtedly identify regulatory factors for lignification in bamboos. Comprehensive investigation of the lignification process using the P. nigra cell culture system in combination with various -omics analyses will provide a valuable framework for accelerating our understanding of the cellular systems regulating lignification in bamboo species.
Materials and Methods
Cell culture
Bamboo (P. nigra) cells were maintained in suspension culture in modified half-strength Murashige and Skoog (MS) liquid medium35 supplemented with 3 μM 2,4-D, as described previously36. Subcultures were established in 100 ml liquid medium in a 300-ml flask and maintained on a rotary shaker (110 rpm) in the dark at 25 °C. To maintain stable morphology and synchronous growth of the cells, the sedimented cell volume was adjusted to 2.5% every two weeks as described previously37.
To promote lignification in the cells, 2-week-old cell cultures were transferred to half-strength MS medium supplemented with 10 μM benzylaminopurine (BA) and 3% (w/v) sucrose (lignification conditions) and cultured as described above12.
Sample preparation for metabolome analysis
The P. nigra cells were immediately frozen in liquid nitrogen and stored at −80 °C until metabolite extraction. Cell samples were weighed and homogenized by Shake Master, BMS-M10N21 (BioMedicalScience, Japan) three times at 1,500 rpm for 2 min, after addition of 500 μl of ice-cold methanol containing 50 μM methionine sulfone as an internal standard. The homogenates were mixed with 500 μl of chloroform and 200 μl of ice-cold Milli-Q water. After centrifugation at 2,300 × g for 5 min at 4 °C, the supernatant was centrifugally filtrated with a Millipore Ultrafree-MC PLHCC HMT Centrifugal Filter Device, 5 kDa (Millipore, Billerica, MA, USA). The filtrate was dried and dissolved in 50 μl of Milli-Q water, and analyzed by CE-TOFMS.
RNA extraction
Total RNA was extracted from P. nigra cells using NucleoSpin RNA (Macherey-Nagel, USA), and quality was checked using an Agilent 2100 Bioanalyzer (Agilent, USA).
Library preparation and sequencing
For Illumina based RNA-sequencing, sequencing libraries were constructed using a TruSeq Sample Preparation Kit (Illumina, Inc.) according to the manufacturer’s instructions. The sequencing libraries were sequenced using a Hiseq2000 sequencer by the paired-end sequencing method for sequences 100 bp in length. For ion torrent based RNA-sequencing, poly(A) + RNAs were purified using the MicroPoly(A)Purist™ Kit (Life Technologies, USA) according to the manufacturer’s instructions. Sequencing libraries were obtained using the Ion Total RNA-Seq Kit v2 (Life Technologies, USA) according to the manufacturer’s instructions with Ion Xpress RNA-Seq Barcode 1–16 Kit (Life Technologies, USA). The sequencing libraries were sequenced using an Ion Proton sequencer by Ion P1 Template OT2 200 Kit v3 (Life Technologies, USA) and Ion P1 Sequencing 200 Kit v3 (Life Technologies, USA).
Read processing
The reads from the Ion Torrent-based sequencing that passed the quality control process of the Ion Torrent system were processed by cutadapt38 to remove sequencing adaptors. The raw reads from the Illumina-based sequencing were trimmed and filtered based on quantity using the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html) with parameter settings of –q 30 –p 80 –v –Q 33.
Reference genome data
The sequence dataset of the draft genome, the coding sequence (CDS), and protein sequences of P. edulis (P. heterocycla var. pubescens) (v1.0) were retrieved from the BambooGDB web site (http://www.bamboogdb.org/)13,39. To generate a dataset of structural gene annotations for the P. edulis genome, we mapped the CDS dataset to the draft genome using the GMAP program with default parameter settings, and estimated exon-intron coordinates for the P. edulis genome. A GFF file of the exon-intron coordinates was used to count reads mapped to each gene using featureCount.
Read Mapping
The Illumina reads were mapped to the P. edulis genome sequence using HISAT240 (version 2.0.5) with default parameter settings. The Ion Torrent reads were mapped to the P. edulis genome sequence using the TMAP program (Life Technologies, USA) (version 3.4.1) with parameter settings of mapall -z -o 2 stage1 map4.
Quantification of gene expression
The featureCounts program (http://bioinf.wehi.edu.au/featureCounts/) was used to compute read counts for each gene annotated in the P. edulis genome and, based on the read counts, the RPM values were calculated.
Identification of differentially expressed genes
Genes showing RPM values ≥1 in at least one sample were defined as expressed genes. Differentially expressed genes (DEGs) were calculated using the DESeq2 program41 running in the R package, with a threshold of adjusted p < 1 × 10−3.
Functional annotation and enrichment test
To predict the functions of P. edulis genes, homology searches were performed using BLASTP (−e = 1e-5, −F = F) against entries of a known protein database (NCBI nr, ftp://ftp.ncbi.nih.gov/blast/db), TIGR Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/), and the protein data present in TAIR release 10 (https://www.arabidopsis.org/). The KAAS web server (http://www.genome.jp/tools/kaas/)42 was used to map the protein sequences of P. edulis to metabolic pathways in the KEGG database. In the KEGG pathway mapping, BLAST was used as a search program and “hsa, dme, cel, ath, sce, cho, eco, nme, hpy, rpr, bsu, lla, cac, mge, mtu, ctr, bbu, syn, bth, dra, aae, mja, ape, osa, gmx, and vvi” as search organisms with the single directional best hit method. The Mercator pipeline (http://mapman.gabipd.org/web/guest)43 in the MapMan web service was used for functional classification of the protein sequences of P. edulis based on MapMan ontology44. The genes putatively encoding transcription factors in P. edulis were annotated based on protein-specific DNA binding domains using Hidden Markov models for 60 transcription factors in plants with an HMMER search45–49. Gene set enrichment analysis (GSEA) of DEGs for MapMan ontology and KEGG pathways was performed by Fisher’s exact test.
CE-TOFMS analysis and data processing
CE-TOFMS analysis was performed using an Agilent CE system combined with a TOFMS (Agilent Technologies, Palo Alto, CA, USA) at Human Metabolome Technologies Inc. (HMT, Japan). The samples were diluted two and five times for cation and anion analysis, respectively. Cationic metabolites were separated through a fused silica capillary (50 μm internal diameter × 80 cm length) with Cation Buffer Solution, H3301-1001 (HMT, Japan). Samples were injected at a pressure of 50 mbar for 10 s with the voltage for the CE set at 27 kV. Electrospray ionization-mass spectrometry (ESI-MS) was conducted in positive-ion mode with voltage set at 3 kV. Anionic metabolites were measured through the fused silica capillary (50 μm internal diameter × 80 cm length) with Anion Buffer Solution, H3302-1021 (HMT, Japan). Samples were injected at a pressure of 50 mbar for 25 s with the voltage for the CE set at 30 kV. The ESI-MS was conducted in the negative-ion mode with the voltage set at 3.5 kV. Mass data for the cationic and anionic metabolites were acquired in a range of 50–1,000 m/z. The data were preprocessed using MasterHands software (HMT, Japan). Each metabolite was identified based on m/z and migration time of the MS peak through database search against the HMT database, and was quantified based on the peak area. Differentially accumulated metabolites were identified with a threshold of p < 0.05 in Welch’s t-test across the sample conditions.
Data Availability
RNA-seq dataset: DDBJ Sequence Read Archive accession number DRA006159.
Electronic supplementary material
Acknowledgements
This work was supported by Grants-in-Aid for Scientific Research (C) (Grant No. 22580387 and 25450519 to S.O.) from the Japan Society for the Promotion of Science (JSPS). The authors also thank the research support person for research staff with family responsibilities in RIKEN to T.U.Y.
Author Contributions
S.O. and K.M. conceived, planned, and supervised the project. S.O. prepared the cell samples. Y.U.-Y. performed the RNA-seq analysis. K.I., T.Y., T.S. and K.M. performed the bioinformatics analysis. T.N., Y.K. and K.S. contributed to biological interpretation of the results. S.O., K.I. and K.M., wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-29645-7.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shinjiro Ogita, Email: ogita@pu-hiroshima.ac.jp.
Keiichi Mochida, Email: keiichi.mochida@riken.jp.
References
- 1.Bystriakova N, Kapos V, Stapleton C, Lysenko I. Bamboo Biodiversity. Unep-Wcmc/Inbar. 2003;1:1–72. [Google Scholar]
- 2.Clark, L. G., Londono, X. & Ruiz-Sanchez, E. In Bamboo: the plant and its uses 1–30, 10.1007/978-3-319-14133-6 (2015).
- 3.Lobovikov, M., Paudel, S., Piazza, M., Ren, H. & Wu, J. World bamboo resources: A thematic study prepared in the framework of the Global Forest Resources, assessment 2005. FAO Tech. Pap. 1–74, http://library.duke.edu/catalog/search/recordid/DUKE004081693 (2007).
- 4.Darabant A, et al. Bamboo biomass yield and feedstock characteristics of energy plantations in Thailand. in. Energy Procedia. 2014;59:134–141. doi: 10.1016/j.egypro.2014.10.359. [DOI] [Google Scholar]
- 5.Kumar R, Chandrashekar N. Fuel properties and combustion characteristics of some promising bamboo species in India. J. For. Res. 2014;25:471–476. doi: 10.1007/s11676-014-0478-6. [DOI] [Google Scholar]
- 6.Gao, J. et al. Characterization of the floral transcriptome of Moso bamboo (Phyllostachys edulis) at different flowering developmental stages by transcriptome sequencing and RNA-seq analysis. PLoS One9 (2014). [DOI] [PMC free article] [PubMed]
- 7.Peng, Z. et al. Transcriptome sequencing and analysis of the fast growing shoots of moso bamboo (Phyllostachys edulis). PLoS One8 (2013). [DOI] [PMC free article] [PubMed]
- 8.Zhang, X. M., Zhao, L., Larson-Rabin, Z., Li, D. Z. & Guo, Z. H. De novo sequencing and characterization of the floral transcriptome of dendrocalamus latiflorus (poaceae: Bambusoideae). PLoS One7 (2012). [DOI] [PMC free article] [PubMed]
- 9.Gamuyao R, et al. Hormone distribution and transcriptome profiles in bamboo shoots provide insights on bamboo stem emergence and growth. Plant Cell Physiol. 2017;58:702–716. doi: 10.1093/pcp/pcx023. [DOI] [PubMed] [Google Scholar]
- 10.Yamada H, et al. Rapid response of Arabidopsis T87 cultured cells to cytokinin through His-to-Asp phosphorelay signal transduction. Biosci. Biotechnol. Biochem. 2004;68:1966–76. doi: 10.1271/bbb.68.1966. [DOI] [PubMed] [Google Scholar]
- 11.Nagata T, Nemoto Y, Hasezawa S. Tobacco BY-2 Cell Line as the “HeLa” Cell in the Cell Biology of Higher Plants. Int. Rev. Cytol. 1992;132:1–30. doi: 10.1016/S0074-7696(08)62452-3. [DOI] [Google Scholar]
- 12.Ogita S, Nomura T, Kishimoto T, Kato Y. A novel xylogenic suspension culture model for exploring lignification in Phyllostachys bamboo. Plant Methods. 2012;8:40. doi: 10.1186/1746-4811-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng Z, et al. The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla) Nat. Genet. 2013;45:456–461. doi: 10.1038/ng.2569. [DOI] [PubMed] [Google Scholar]
- 14.Wysocki WP, Ruiz-Sanchez E, Yin Y, Duvall MR. The floral transcriptomes of four bamboo species (Bambusoideae; Poaceae): support for common ancestry among woody bamboos. BMC Genomics. 2016;17:384. doi: 10.1186/s12864-016-2707-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lahens NF, et al. A comparison of Illumina and Ion Torrent sequencing platforms in the context of differential gene expression. BMC Genomics. 2017;18:602. doi: 10.1186/s12864-017-4011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schuetz M, et al. Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiol. 2014;166:798–807. doi: 10.1104/pp.114.245597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, et al. LACCASE 5 is required for lignification of the Brachypodium distachyon culm. Plant Physiol. 2015;168:192–204. doi: 10.1104/pp.114.255489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tohge T, Watanabe M, Hoefgen R, Fernie AR. Shikimate and Phenylalanine Biosynthesis in the Green Lineage. Front. Plant Sci. 2013;4:1–13. doi: 10.3389/fpls.2013.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Chantreau M, Sibout R, Hawkins S. Plant cell wall lignification and monolignol metabolism. Front. Plant Sci. 2013;4:220. doi: 10.3389/fpls.2013.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alejandro S, et al. AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr. Biol. 2012;22:1207–12. doi: 10.1016/j.cub.2012.04.064. [DOI] [PubMed] [Google Scholar]
- 21.Sibout R, Höfte H. Plant Cell Biology: The ABC of Monolignol Transport. Curr. Biol. 2012;22:R533–R535. doi: 10.1016/j.cub.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 22.Takeuchi M, Kegasa T, Watanabe A, Tamura M, Tsutsumi Y. Expression analysis of transporter genes for screening candidate monolignol transporters using Arabidopsis thaliana cell suspensions during tracheary element differentiation. J. Plant Res. 2018;131:297–305. doi: 10.1007/s10265-017-0979-4. [DOI] [PubMed] [Google Scholar]
- 23.Taylor-Teeples M, et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature. 2015;517:571–5. doi: 10.1038/nature14099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell. 2008;20:2763–2782. doi: 10.1105/tpc.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Q, Dixon RA. Transcriptional networks for lignin biosynthesis: More complex than we thought? Trends in Plant Science. 2011;16:227–233. doi: 10.1016/j.tplants.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Hirano K, et al. Identification of transcription factors involved in rice secondary cell wall formation. Plant Cell Physiol. 2013;54:1791–1802. doi: 10.1093/pcp/pct122. [DOI] [PubMed] [Google Scholar]
- 27.Lin Z, Zhong S, Grierson D. Recent advances in ethylene research. J. Exp. Bot. 2009;60:3311–3336. doi: 10.1093/jxb/erp204. [DOI] [PubMed] [Google Scholar]
- 28.Lin RC, Park HJ, Wang HY. Role of Arabidopsis RAP2.4 in regulating lightand ethylene-mediated developmental processes and drought stress tolerance. Mol. Plant. 2008;1:42–57. doi: 10.1093/mp/ssm004. [DOI] [PubMed] [Google Scholar]
- 29.Pesquet E, Tuominen H. Ethylene stimulates tracheary element differentiation in Zinnia elegans cell cultures. New Phytol. 2011;190:138–149. doi: 10.1111/j.1469-8137.2010.03600.x. [DOI] [PubMed] [Google Scholar]
- 30.Cook CM, et al. Transcriptional changes related to secondary wall formation in xylem of transgenic lines of tobacco altered for lignin or xylan content which show improved saccharification. Phytochemistry. 2012;74:79–89. doi: 10.1016/j.phytochem.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zdarska M, et al. Illuminating light, cytokinin, and ethylene signalling crosstalk in plant development. In. Journal of Experimental Botany. 2015;66:4913–4931. doi: 10.1093/jxb/erv261. [DOI] [PubMed] [Google Scholar]
- 32.Van de Poel B, Smet D, Van Der Straeten D. Ethylene and Hormonal Cross Talk in Vegetative Growth and Development. Plant Physiol. 2015;169:61–72. doi: 10.1104/pp.15.00724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, Zhang H, Ai Q, Liang G, Yu D. Two bHLH Transcription Factors, bHLH34 and bHLH104, Regulate Iron Homeostasis in Arabidopsis thaliana. Plant Physiol. 2016;170:2478–93. doi: 10.1104/pp.15.01827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar M, Campbell L, Turner S. Secondary cell walls: Biosynthesis and manipulation. Journal of Experimental Botany. 2016;67:515–531. doi: 10.1093/jxb/erv533. [DOI] [PubMed] [Google Scholar]
- 35.Murashige T, Skoog F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 36.Ogita S. Callus and cell suspension culture of bamboo plant, Phyllostachys nigra. Plant Biotechnol. 2005;22:119–125. doi: 10.5511/plantbiotechnology.22.119. [DOI] [Google Scholar]
- 37.Ogita S, Kikuchi N, Nomura T, Kato Y. A practical protocol for particle bombardment-mediated transformation of phyllostachys bamboo suspension cells. Plant Biotechnol. 2011;28:43–50. doi: 10.5511/plantbiotechnology.10.1101a. [DOI] [Google Scholar]
- 38.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 39.Zhao, H. et al. BambooGDB: A bamboo genome database with functional annotation and an analysis platform. Database2014 (2014). [DOI] [PMC free article] [PubMed]
- 40.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Love, M. I., Anders, S. & Huber, W. Differential analysis of count data - the DESeq. 2 package. Genome Biology15 (2014).
- 42.Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C. & Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35 (2007). [DOI] [PMC free article] [PubMed]
- 43.Lohse M, et al. Mercator: A fast and simple web server for genome scale functional annotation of plant sequence data. Plant, Cell Environ. 2014;37:1250–1258. doi: 10.1111/pce.12231. [DOI] [PubMed] [Google Scholar]
- 44.Thimm O, et al. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313X.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 45.Tran, L.-S. P. & Mochida, K. Identification and prediction of abiotic stress responsive transcription factors involved in abiotic stress signaling in soybean. Plant Signal. Behav. 5 (2010). [DOI] [PMC free article] [PubMed]
- 46.Mochida, K. et al. In silico analysis of transcription factor repertoires and prediction of stress-responsive transcription factors from six major gramineae plants. DNA Res. 18 (2011). [DOI] [PMC free article] [PubMed]
- 47.Mochida, K. et al. LegumeTFDB: An integrative database of Glycine max, Lotus japonicus and Medicago truncatula transcription factors. Bioinformatics26 (2010). [DOI] [PubMed]
- 48.Mochida K, et al. In silico analysis of transcription factor repertoire and prediction of stress responsive transcription factors in soybean. DNA Res. 2009;16:353–369. doi: 10.1093/dnares/dsp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mochida, K. et al. TreeTFDB: An integrative database of the transcription factors from six economically important tree crops for functional predictions and comparative and functional genomics. DNA Res. 20 (2013). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq dataset: DDBJ Sequence Read Archive accession number DRA006159.