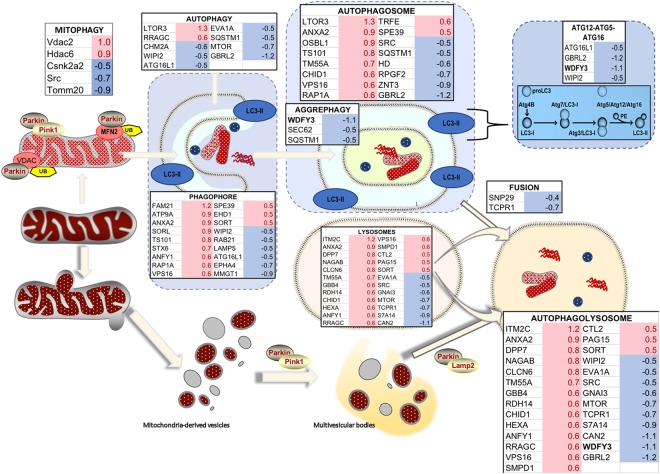
Figure 9.
Differential expression of proteins involved in autophagy and mitophagy in brain of WT and Wdfy3 mice. Proteomics was performed on cortical mitochondria-enriched fractions from WT and Wdfy3+/lacZ mice (n = 7/genotype). Proteomics data were filtered as described in the Methods and main manuscript sections. The fold changes for these proteins were calculated by using the log 2 of the value for haploinsufficient over WT for each feature. Only those with 0.5 ≤ log2 FC ≤ −0.5 are shown. Upon Pink1-dependent recruitment of Parkin to depolarized mitochondria (mitophagy), the defective organelles are internalized within the autophagosome and degraded upon fusion of the autophagosome with lysosomes. Proteins involved in aggrephagy (to clear damaged, unfolded proteins shown as red, wavy structures), as well as ATG12-ATG5-ATG16l complex, were present at lower levels in mitochondria-enriched fractions from cortex of Wdfy3-haploinsufficient mice vs. WT. However, proteins involved in phagophore and autophagosome formation seemed enriched in these fractions (~2-fold), but accompanied by a decreased level of proteins mediating the fusion of autophagosomes with lysosomes. An autophagic independent degradation pathway exists, through the formation of mitochondria-derived vesicles and subsequent fusion with lysosomes. In red are shown upregulated proteins, in blue downregulated ones.