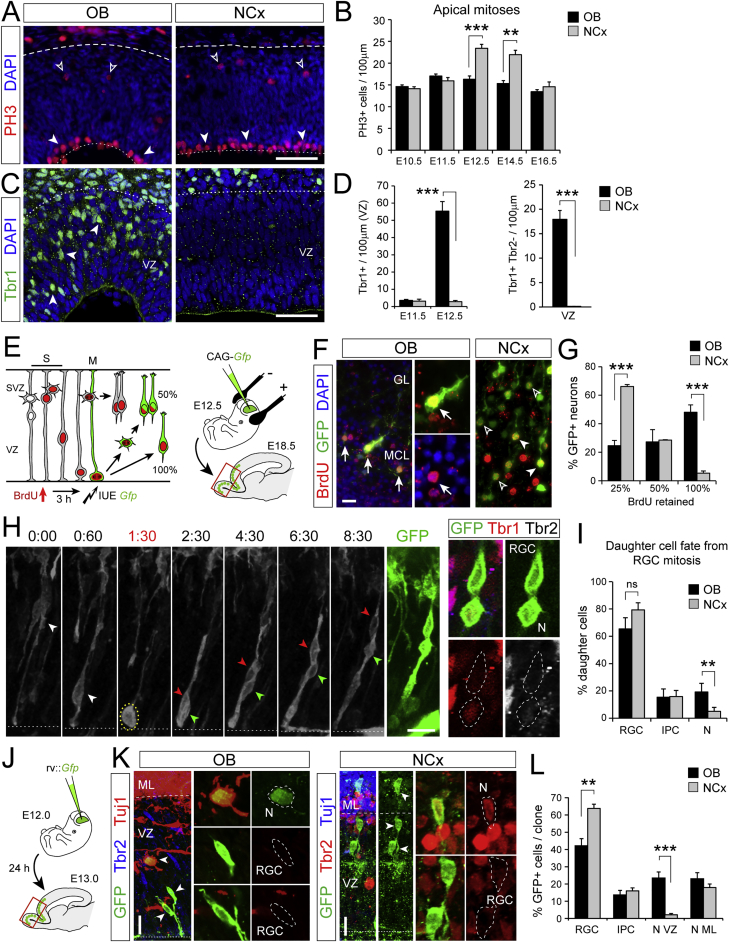
Figure 2.
Higher Frequency of Direct Neurogenesis in OB than NCx
(A–D) Distribution of apical (solid arrowheads) and basal (open arrowheads) mitoses (A) and neurons (C) at E12.5, and quantifications as indicated (n = 3–11 embryos per group; t tests).
(E–G) Experimental design to identify neurons born by direct or indirect neurogenesis based on GFP labeling and BrdU retention, representative examples (OB, mitral cells with 100% BrdU [arrows]; NCx, projection neurons with 25% and 50% BrdU) and quantifications (n = 4 embryos per group; t tests).
(H and I) Time-lapse imaging frames from an RGC dividing in the OB (t = 1:30), and analysis of daughter cells (n = 44 divisions in NCx, 25 divisions in OB; 3 and 5 embryos, respectively; χ2-test).
(J–L) Experimental design for clonal analysis, representative examples and quantification (n = 116 clones NCx, 47 clones OB, 7 embryos; χ2-test). Arrowheads indicate soma of cells in clone. GL, glomerular layer; MCL, mitral cell layer.
Values are mean + SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Scale bars: 50 μm (A and C), 10 μm (F, H, and K).
See also Figures S2 and S3.