Figure S7.
Robo/Dll1 Signaling Drives Direct Genesis of Deep-Layer Corticofugal Neurons and Regulates Direct Neurogenesis in Brain Regions with Ancestral Origin, Related to Figure 5
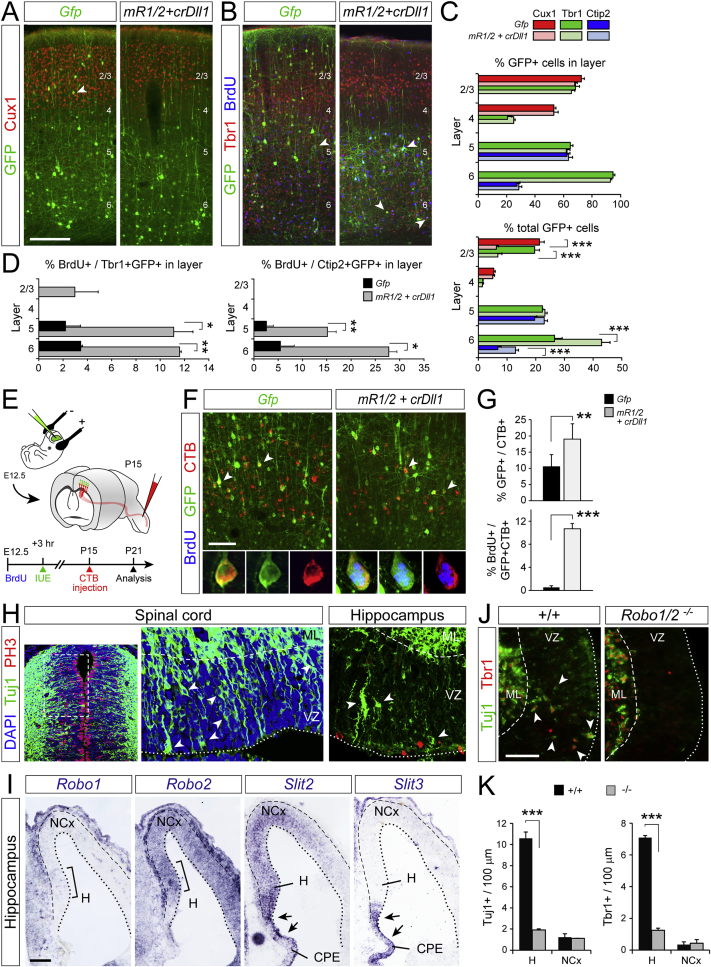
(A–D) Analysis of fate marker expression by directly generated neurons (arrowheads) upon electroporation with the indicated plasmid combinations. Plots in (C) show the proportion of GFP+ cells within each layer (top) and of all GFP+ cells in NCx (bottom) expressing each marker. Plots in (D) show the proportion of cells positive for GFP and Tbr1 (left), or Ctip2 (right), within each layer that retain 100% BrdU label (n = 3-5 animals per condition; one way ANOVA followed by χ2-test).
(E) Experimental design to determine axonal projection identity of neocortical neurons born by direct neurogenesis at E12.5.
(F and G) Retrograde labeling of corticospinal-projecting neurons generated directly upon electroporation with the indicated plasmid combinations, and analysis of abundance. Insets show soma of single CTB+GFP+ neurons. Plots in (G) show proportion of CTB-traced neurons expressing GFP (top; arrowheads in (F) and proportion of CTB+GFP+ neurons retaining 100% BrdU label (bottom) (n = 3-4 animals per condition; χ2-test).
(H) Tuj1 and PH3 stains of the embryonic spinal cord and hippocampal primordium, showing that virtually all mitoses are apical and neurons are very abundant in the VZ of both regions, two traits indicative of direct neurogenesis.
(I) ISH in coronal sections of hippocampus and adjacent neocortex at E12.5. Expression of Robo1, Robo2, and of Slit2 and Slit3, mRNAs is most prominent in hippocampus primordium (H) and choroid plexus epithelium (CPE), respectively.
(J and K) Distribution and abundance of Tuj1+ / Tbr1+ neurons (arrowheads) in hippocampal primordium and adjacent neocortex from control and Robo1/2−/− mutants at E12.5 (n = 3 embryos per group; t tests).
Values are mean + SEM; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗p < 0.001. Scale bars: 200 μm (A and B); 100 μm (I), 50 μm (F and J).