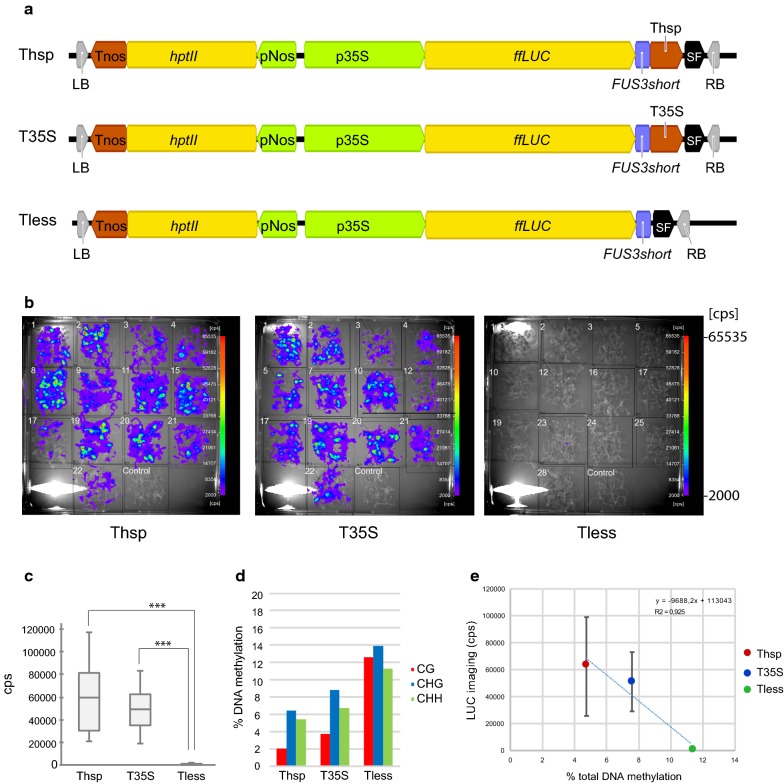
Fig. 1.
Luciferase activity and DNA methylation analysis of the 35S CaMV promoter in 10 day-old seedlings (d.o.s). a Schematic representation of the constructs used to transform Arabidopsis. b Luciferase imaging in 10 d.o.s. of T2 generation measured as counts per seconds (cps). c Box plot showing luciferase activity measures, represented as cps per line. *** represents highly significant differences (p < 0.001) according to Student’s test. d DNA methylation quantification of CG (red bars), CHG (blue bars) and CHH (green bars) cytosine contexts. e Luciferase activity ± standard deviation versus total DNA methylation plot for 10 d.o.s. % of DNA methylation was calculated as (number of methylated C residues in each context (CG, CHG or CHH)/total number of C residues in that context) * 100. % of total DNA methylation was calculated as (number of methylated C residues/total number of C residues) * 100. LB left border, RB right border, Tnos nopaline synthase terminator, hptII hygromycin resistance, pNos nopaline synthase promoter, p35S 35S CaMV promoter, ffLUC firefly luciferase, FUS3short FUS3 PEST sequence that includes aa 246–283, Thsp heat shock protein 18.2 terminator, T35S 35S CaMV terminator, SF stuffer fragment