Fig. 3.
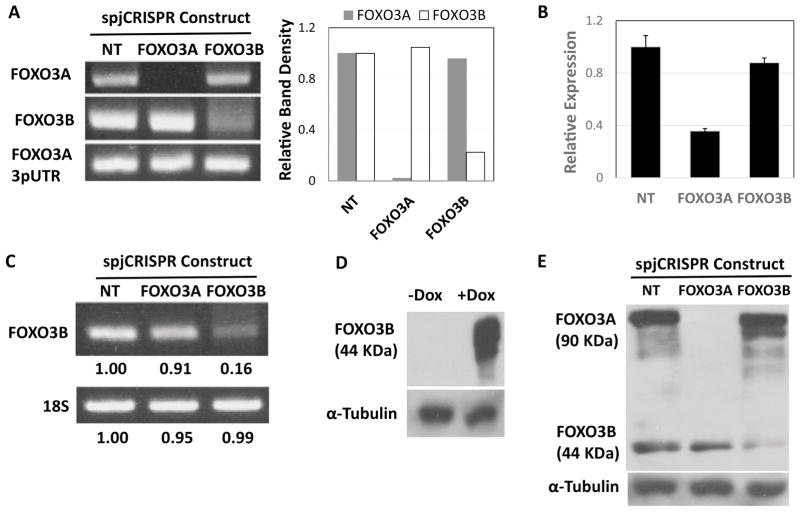
spJCRISPR achieves specific knockdown of FOXO3A and FOXO3B on the mRNA and protein level. A. Genomic PCR using primers flanking the FOXO3A and FOXO3B target sites for the respective spJCRISPR constructs. LHCNM2 myoblasts were infected with the described constructs and selected on puromycin. A week following selection genomic DNA was harvested and PCR performed (35 cycles). Primers specific to sequence in the FOXO3A 3pUTR were used as a loading control. Band densitometry was done by normalizing all bands to their respective NT construct band intensities. Primer pair locations are annotated in Figure 2A. B. RT-qPCR of FOXO3A mRNA using the FOXO3A-specific 3pUTR primers from RNA harvested in parallel with the gDNA. FOXO3A mRNA measurements were normalized to 18S rRNA and calculated relative to cells infected with the NT construct using the ΔΔCt method. Errors bars are SD of four replicates. Primer locations are annotated in Figure 2A. C. RT-PCR of FOXO3B mRNA using FOXO3B-specific primers and 18S rRNA primers as a loading control from the RNA harvested in parallel with the gDNA (35 cycles). Band densitometry was done as in Figure 3A. Primer locations are annotated in Figure 2A. D. Overexpression of FOXO3B in LHCNM2 myoblasts infected with a Dox-inducible FOXO3B expression construct. +Dox cells were treated with 0.1 ug/ml Dox for 48 hours and protein harvested. The western blot was performed with a monoclonal FOXO3A antibody raised against FOXO3A n-terminal residues with the epitope centered on Glu50 (Cell Signaling #2497). α-Tubulin is blotted as a loading control. E. Western blot of endogenous FOXO3A and FOXO3B in LHCNM2 myoblasts from protein harvested in parallel with the RNA and gDNA. α-Tubulin is used as the loading control.