Fig. 3.
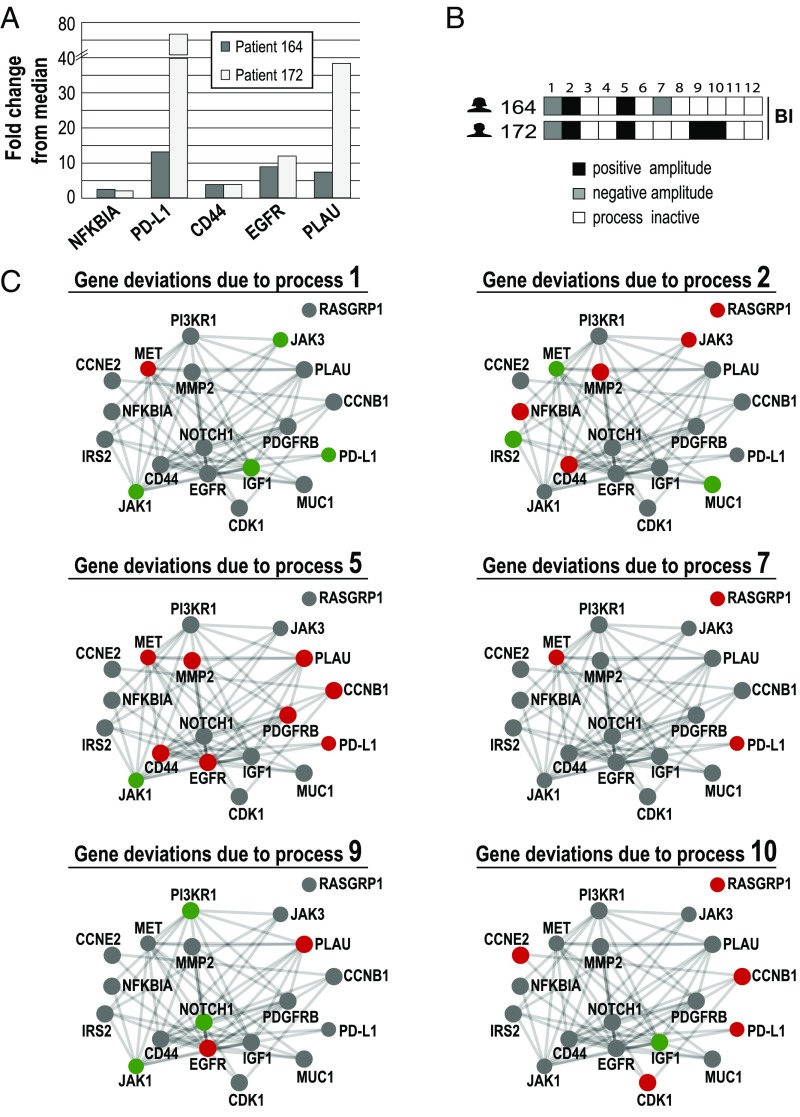
Similar gene expression levels in different patients may be attributed to different unbalanced processes. Two bladder cancer (Bl) patients were selected for the demonstration of the concept. (A) The fold changes of five selected Bl-associated oncogenes are shown. NF-κBIA, PD-L1, CD44, EGFR, and PLAU were up-regulated in both patients relative to their median expression levels across 506 patients. (B) Patient-specific combinations of unbalanced processes. Patient 164 harbors processes 1, 2, 5, and 7. Patient 172 harbors unbalanced processes 1, 2, 5, 9, and 10. Selected genes from these processes are shown in C. (C) The influences of these unbalanced processes on 19 selected oncogenes are shown. The complete list of transcripts that participate in each of the processes (∼370–1,500 transcripts for each process) can be found in Dataset S1. Red denotes up-regulation, green denotes down-regulation, and gray denotes no change due to the process. Functional connections are according to STRING database. For example, CD44 was up-regulated in both patients to a similar extent. This is attributed to unbalanced processes 2 and 5, which are active in both patients. The up-regulation of PD-L1 in patient 164 is attributed to processes 5 and 7, whereas in patient 172, the up-regulation in PD-L1 is attributed to processes 5 and 10. The up-regulation of PLAU in both patients is associated with process 5. However, in patient 172, the up-regulation of PLAU is also attributed to process 9, which is active in his tumor as well.