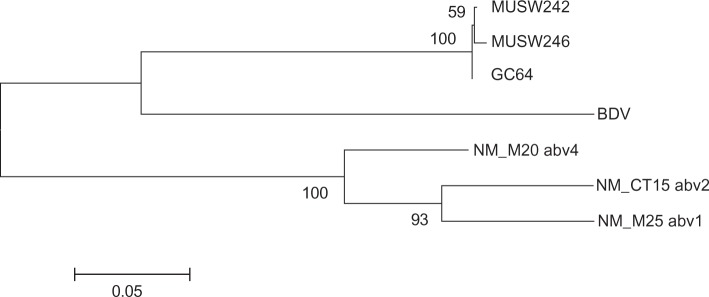
Figure 1.
Phylogeny generated using an 1100 nt fragment of bird and mammal bornaviruses that includes part of the N gene, the N/X intergenic region, the entire X gene, and part of the P gene.
Notes: The evolutionary history was inferred using the Maximum Likelihood method based on the HKY model.11 A bootstrap consensus tree was inferred from 1000 replicates.12 The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved seven nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 980 positions in the final dataset. Evolutionary analyses were conducted in MEGA5.13 Accession numbers are: NC_001607 (BDV), GU249595.2 (ABV1 NM-M25), HM998710 (ABV2 NM-CT15), JN014949 (ABV4 NM-M20), JQ687270(GJ_MUSW242_2012) and JQ687271 (GJ_MUSW246_2012).