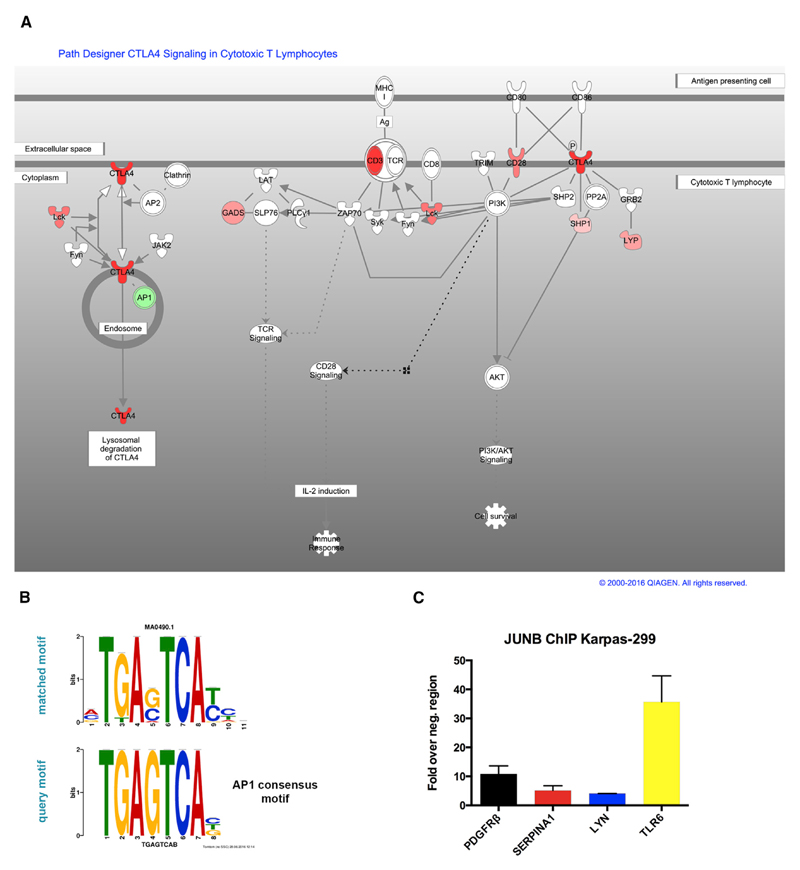
Figure 6. Canonical Pathways Are Affected by Differentially Methylated Genes.
(A) Multiple genes of the TCR pathway are hypermethylated in ALK+ and ALK− ALCL (red, significantly hypermethylated genes; green, significantly hypomethylated genes; adjusted p value < 0.05; β-value difference ≥ 0.15). The pathway was generated through the use of QIAGEN’s IPA.
(B) DNA sequence motif identified by unbiased motif search in regions adjacent to hypomethylated CpGs in ALK+ and ALK− ALCL (top) compared to the AP1 consensus motif (bottom).
(C) ChIP for JUNB occupancy at hypomethylated ALCL promoters with putative AP1 binding sites. PDGFRβ, positive control. ChIP was normalized to a negative control region in the 3′ end of the PDGFRβ; gene containing no AP1 motif. Values are means ± SEM. Each value is the mean of three replicates.
See also Figures S5–S8.