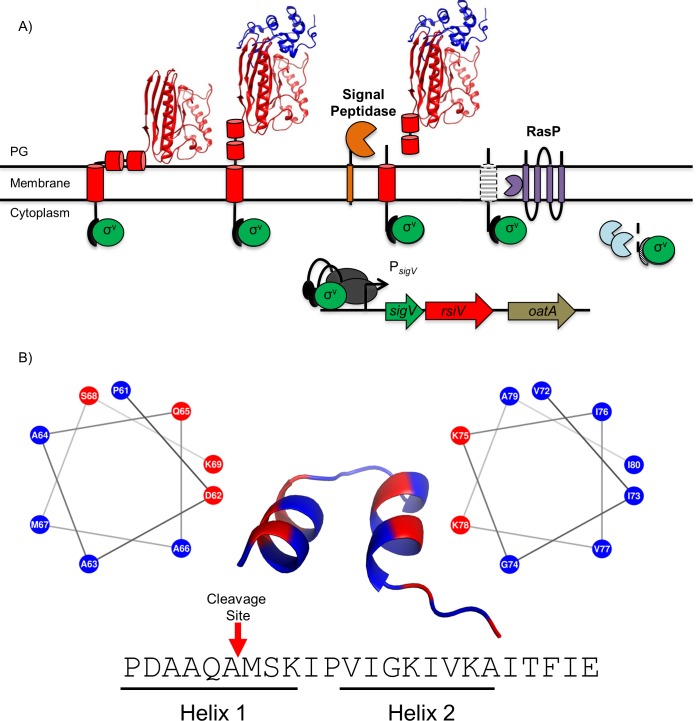
Fig 1. Model of σV activation.
A. The σ factor σV is shown in green, the anti-σ factor RsiV is shown in red with cartoon cylinders representing the unsolved residues 1–75. Signal peptidase (Sip) is shown in yellow, the site two protease RasP is shown in purple and lysozyme is shown in blue. In the absence of lysozyme, RsiV is resistant to signal peptidase cleavage via the interaction of two amphipathic helices with the membrane that restrict access to the cleavage site. In this state RsiV sequesters σV activity and prevents transcription of σV-dependent genes. Once RsiV binds lysozyme, the amphipathic helices are forced out of the membrane and signal peptidase is allowed access to the cleavage site leading to site-1 degradation of RsiV. The remaining transmembrane portion of RsiV is further degraded by the site-2 protease RasP, after which RsiV is further degraded by cytosolic proteases. This allows σV to interact with RNA polymerase (Grey) and promote transcription of σV-dependent genes. B. An set of 203 RsiV homologs were aligned using Clustal Omega [70] and the alignment was used with weblogo [44] to generate a sequence logo (S1 Fig). The corresponding sequence in B. subtilis was run through the secondary structure prediction software PEP-FOLD-3 [45] and the results are shown by the cartoon structure with two α-helices. Each helix was modeled with a helical wheel generator [47] and labeled below the sequence designated as Helix 1 (Left) and Helix 2 (Right). Hydrophobic amino acids are represented in blue and hydrophilic residues are represented in red. The cleavage site of RsiV is represented with a red arrow. This bioinformatic analysis suggests the presence of two conserved amphipathic helices directly after the transmembrane domain of RsiV that function to protect RsiV from site-1 cleavage by signal peptidase.