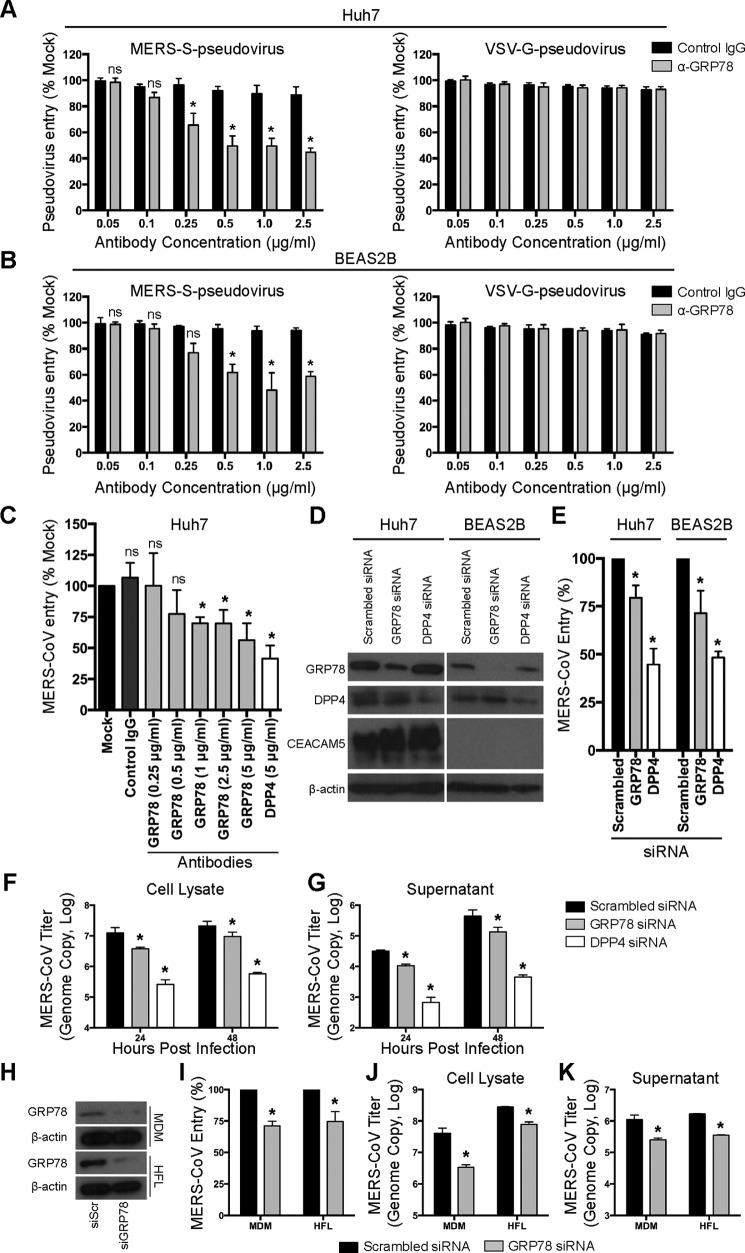
Figure 5.
GRP78 is involved in MERS-CoV entry. Pseudovirus antibody blocking assays were performed in Huh7 (A) and BEAS2B (B) cells. A titration of GRP78 or isotype control antibodies from 0 to 2.5 μg/ml was added and pre-incubated with Huh7 and BEAS2B cells for 1 h at 37 °C. MERS–S-pseudovirus or VSV-G–pseudovirus was subsequently added at a ratio of 100 LP per cell for 1 h. Luciferase activity was determined at 72 h post-inoculation and was normalized to that of the mock-treated cells. C, antibody blocking assay was performed in Huh7 cells using infectious MERS-CoV. Huh7 cells were pre-incubated with antibodies at the indicated concentration for 1 h at 37 °C. The cells were then challenged with MERS-CoV at 1 m.o.i. for 1 h at 37 °C in the presence of the antibodies. After 1 h, the cells were washed and harvested. MERS-CoV entry was assessed with qPCR, and the result was normalized to that of the mock-treated cells. D, Huh7 or BEAS2B cells were treated with 75 nm GRP78, DPP4, or scrambled siRNA for 2 consecutive days. The knockdown efficiency was evaluated with Western blottings. E, siRNA-treated Huh7 or BEAS2B cells were infected with MERS-CoV at 1 m.o.i. for 1 h at 37 °C. After 1 h, the cells were harvested, and virus entry was evaluated with qPCR analysis. The result was normalized to that of the scrambled siRNA-treated cells. siRNA-treated BEAS2B cells were infected with MERS-CoV at 0.1 m.o.i. for 1 h at 37 °C. The cell lysates (F) and supernatants (G) were harvested at 24 and 48 h post-infection. MERS-CoV replication was evaluated with qPCR analysis. H, siRNA-treated MDM or HFL was infected with MERS-CoV at 1 m.o.i. for 2 h at 37 °C. After 2 h, the cells were harvested, and virus entry was evaluated with qPCR analysis (I). The result was normalized to that of the scrambled siRNA-treated cells. siRNA-treated MDM or HFL was infected with MERS-CoV at 0.1 m.o.i. for 1 h at 37 °C. The cell lysates (J) and supernatants (K) were harvested at 24 h post-infection. MERS-CoV replication was evaluated with qPCR analysis. In all panels, data represented mean and S.D. from three independent experiments. Statistical analyses were carried out using Student's t test. Statistical significance was indicated by asterisks when p < 0.05. ns means not significant.