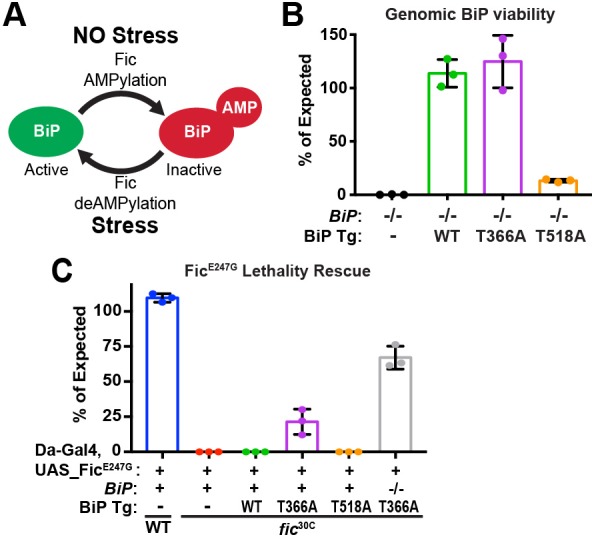
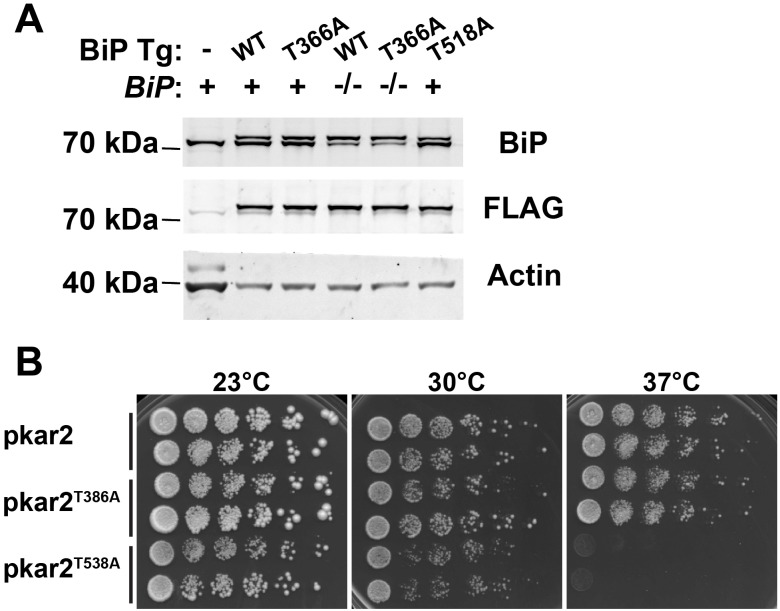
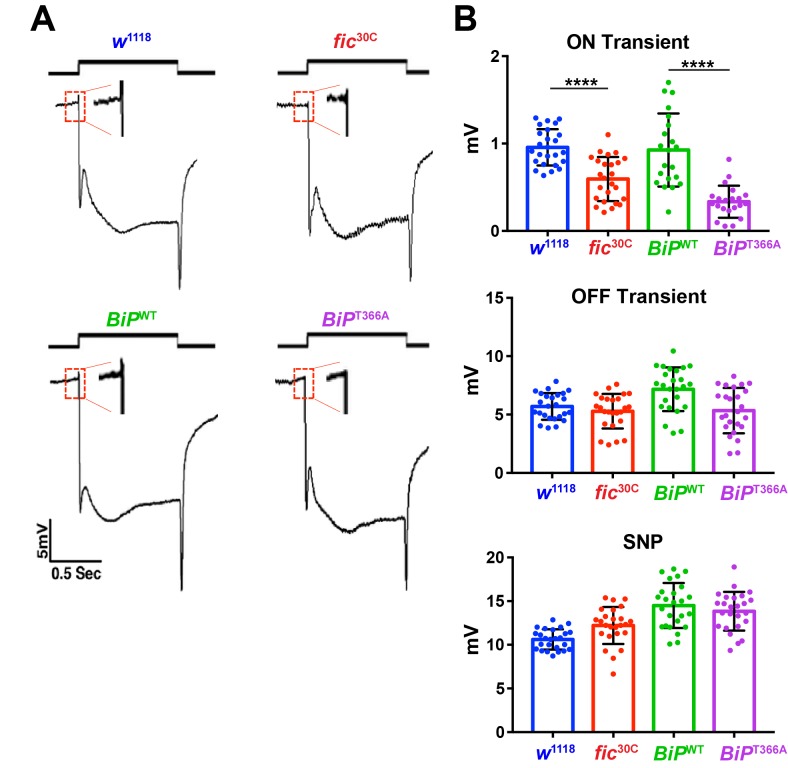
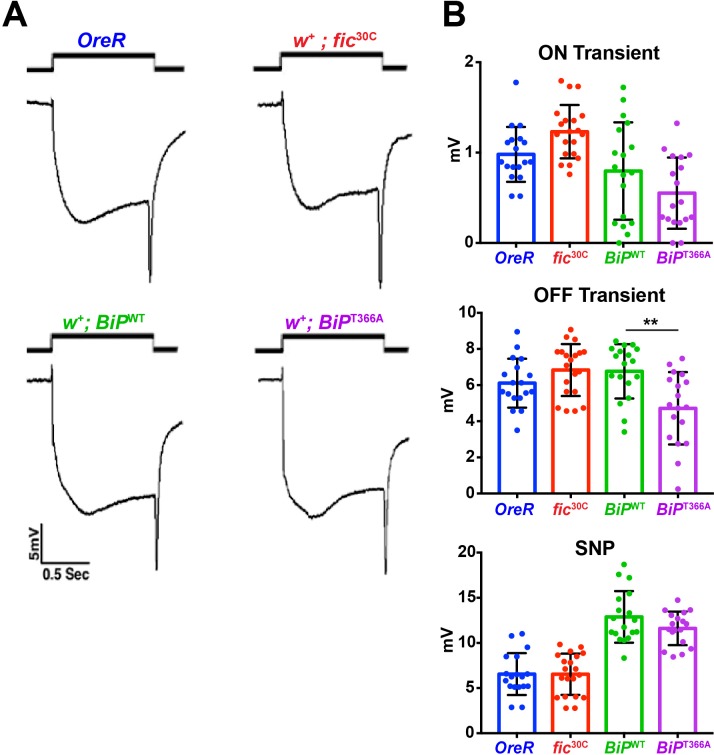
Figure 1. BiP is a target of Fic AMPylation and deAMPylation in vivo.
(A) BiP AMPylation during times of low ER stress reserves a portion of the chaperone to allow for a rapid, deAMPylation-driven, response to high ER stress (Casey et al., 2017; Preissler et al., 2017a). (B) Bar graphs show the percentage of null mutant BiPG0102/y males rescued by the indicated genomic BiPWT, BiPT366A or BiPT518A genomic transgene (Tg) relative to sibling controls. N = 3 biological replicas. At least 50 flies scored for each replica. Bar graphs show means ± Standard Deviation (SD). (C) Bar graphs show the percentage of viable flies of the indicated wild-type or fic30C genotypes expressing the overactive FicE247G under the ubiquitous Da-Gal4 driver relative to sibling controls. Among the indicated genomic BiP transgenes, only BiPT366A provides partial rescue of lethality in the BiP+/+ background and near complete rescue in a BiPG0102 null background. N = 3 biological replicas. At least 100 total flies scored for each replica. Bar graphs show means ± SD.