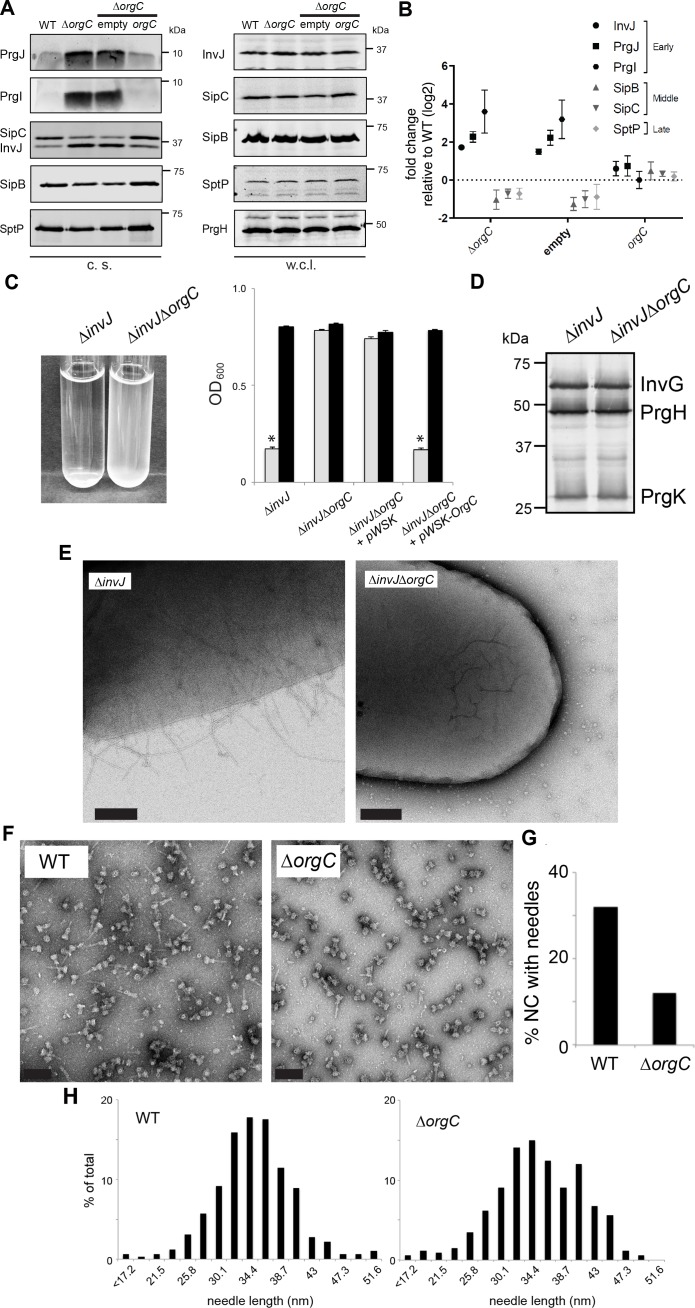
Figure 2. OrgC is necessary for efficient needle assembly but does not affect needle length.
(A) S. Typhimurium ΔorgC secretes elevated amount of early and reduced amount of middle/late substrates. Proteins in bacterial culture supernatants (c. s.) were concentrated by TCA precipitation and analyzed by SDS-PAGE, followed by immunoblot using specific antibodies for PrgI, PrgJ, InvJ (early substrates), SipB, SipC (middle substrates) and SptP (late substrate). The secretion phenotype of ΔorgC was complemented by introducing a plasmid expressing OrgC but not by introducing the empty plasmid vector alone. Expression levels of the indicated proteins in whole-cell lysates (w. c. l.) were also evaluated. (B) Western-blot band intensities from three independent experiments were quantified. Values were normalized to the wild-type parent strain and log2 transformed. The dotted horizontal line corresponds to the levels of the different proteins in wild type S. Typhimurium. (C) A bacterial cell-clumping assay implicates OrgC in needle assembly. Cultures of the S. Typhimurium ∆invJ mutant strain, which display long needle filaments, clump and precipitate to the bottom of the tube (left panel) resulting in drastically decreased OD600 (right panel, grey bars), which can be recovered by vortexing the samples (right panel, black bars). Bacterial cell clumping is abolished by introduction of a ∆orgC mutation (left and right panels), which can be complemented by introducing a plasmid encoding orgC (pWSK-orgC) but not by introducing the vector alone (pWSK) (right panel). Values represent OD600 before (grey bars) and after (black bars) vortexing and are the mean ± SEM (standard error of the mean) of three independent measurements. Asterisks indicate statistically significant differences from the values of the vortexed sample (p<0.001, Student t test). (D) Western blot analysis of the NC base components InvG, PrgH, and PrgK in whole cell lysates of the ∆invJ and ∆invJ ∆orgC S. Typhimurium mutant strains. (E) Electron micrographs of negatively stained S. Typhimurium showing the presence (∆invJ) or absence (∆invJ ∆orgC) of long T3SS needle filaments. Scale bar = 200 nm, (∆invJ); 100 nm (∆invJ ∆orgC). (F) Electron micrographs of negatively stained needle complexes isolated from wild type (WT) or ∆orgC S. Typhimurium. Scale bar = 100 nm. (G) Percentage of needle complexes exhibiting the needle filament in preparations obtained from wild type (WT) or ∆orgC S. Typhimurium (number of particles analyzed: w. t. = 1105; ∆orgC = 1273). (H) Needle length of needle complexes isolated from S. Typhimurium wild type or ∆orgC mutant strains. The percentage of needle complexes exhibiting the indicated length (in nm, x axis) is indicated (number of needle complexes analyzed: w. t. = 314; ∆orgC = 339.
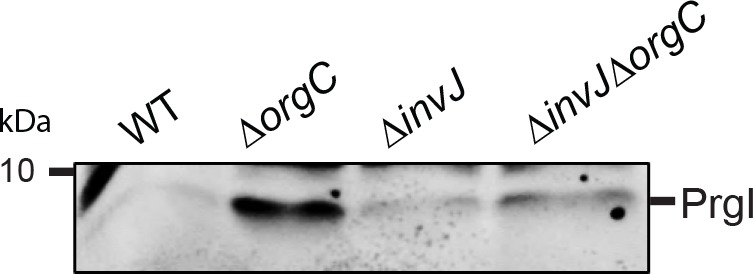
Figure 2—figure supplement 1. PrgI secretion profile of S.