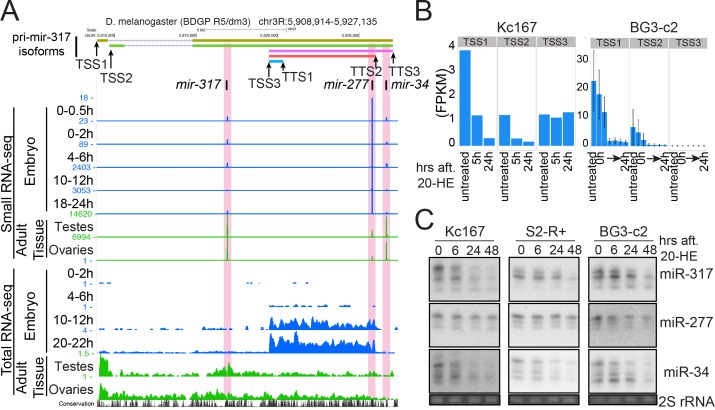
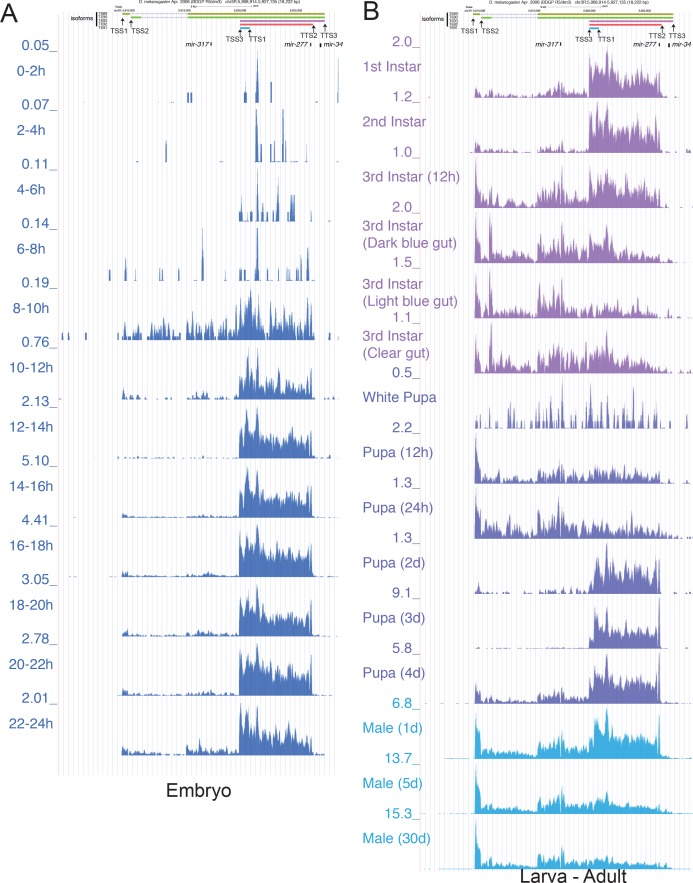
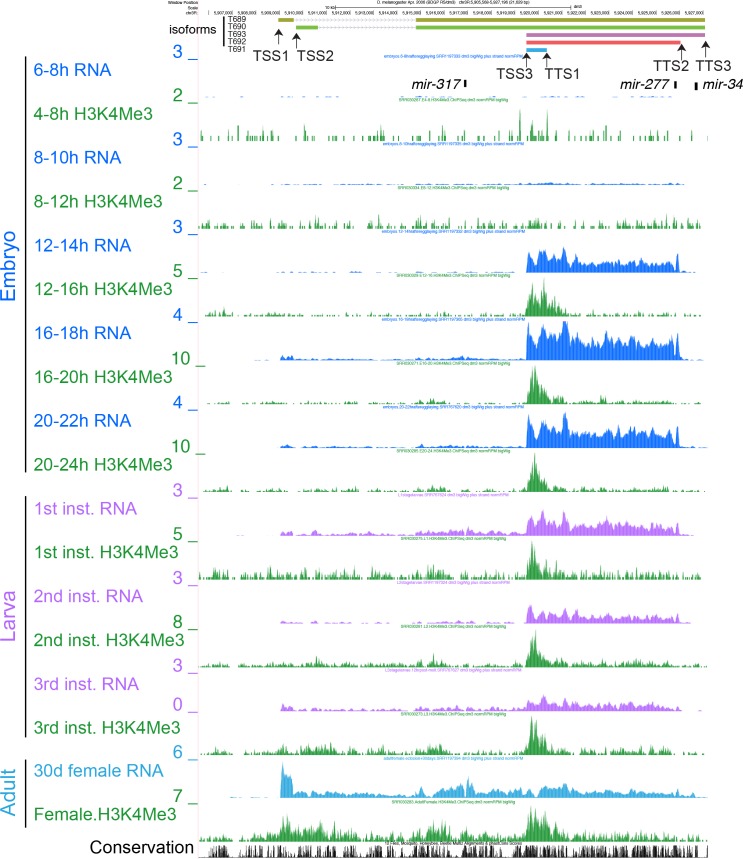
Figure 5. Distinct miRNA subsets are produced from the mir-317 cluster by alternative pri-miRNA isoforms.
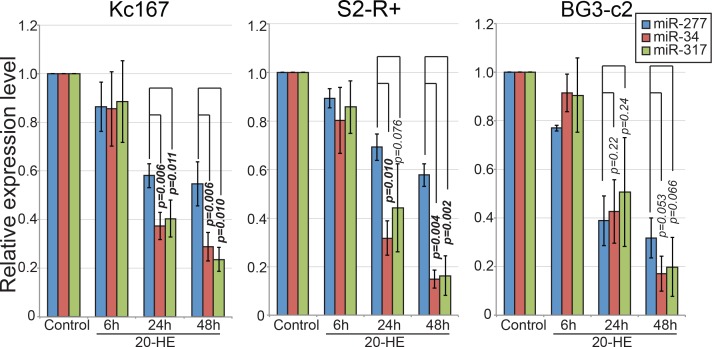
(A) UCSC genome browser snapshot of the mir-317 cluster locus. Small RNA-seq (upper) and total RNA-seq (lower) data are shown. In embryos (blue) miR-277 expression is higher than the other two miRNAs, and even higher in late embryos, which is consistent with the high expression of pri-miRNA isoform starting from TSS3 in embryos. In testes and ovaries, long isoforms starting from TSS1 and 2 are dominant, and the miR-277 level is lower than miR-317 and miR-34. In all tissues and embryonic stages, the ratios between miR-317 and miR-34 remain similar. (B) Reanalysis of published total RNA-seq data from cultured cells treated with ecdysone. The sum of FPKMs of the isoforms sharing the same TSS was plotted. (C) Northern blotting analysis of mature miR-317,–277 and −34 levels after 20-HE (hydroxyecdysone) addition in Kc167, S2-R+ and BG3-c2 cells. Cells were treated with 20-HE for the indicated time and total RNA was separated on a 15% denaturing acrylamide gel. miR-317 and miR-34 were decreased after 20-HE addition in Kc167 and S2-R+ cells, while the decrease of miR-277 was much weaker in these cell lines. In contrast, miR-277 was also decreased in BG3-c2 cells, in which the level of the mir-277 specific short isoform was very low even in the absence of 20-HE (Panel B, TSS3). See Figure 5—figure supplement 3for the quantified results of tripricates.