FIG 6.
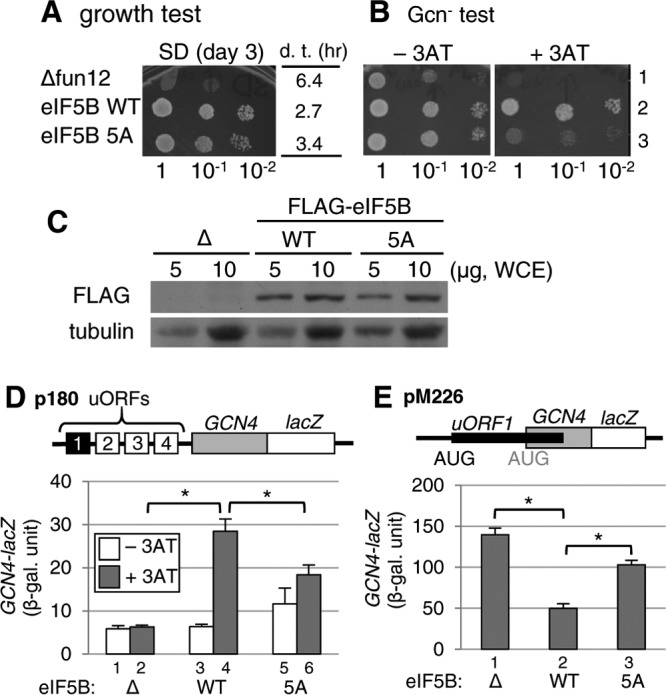
Effects of the eIF5B-5A mutation that disrupts the eIF5B·stalk interaction in yeast. (A and B) Growth and Gcn− phenotype tests. Fixed amounts (5 μl of culture diluted to an OD600 of 0.15) of Δfun12 strain J111 carrying the control vector (Δfun12) (row 1) or the single-copy plasmid pC1818 expressing FLAG-tagged eIF5B-WT (row 2) or eIF5B-5A (row 3) and their 10-fold serial dilutions were spotted onto synthetic dextrose (SD) (A) or SC (B) medium lacking histidine, with or without 4 mM 3AT, and then incubated for 3, 2, or 5 days at 30°C, respectively. In panel A, the table indicates the doubling time (d. t.) of each transformant in liquid medium. (C) Expression of plasmid-directed wild-type eIF5B and the eIF5B-5A mutant in yeast Δfun12 cells. Whole-cell extracts (WCE) from the Δfun12 strain and the same strains transformed with wild-type eIF5B and eIF5B-5A mutant genes with FLAG tags were subjected to immunoblot analyses using anti-FLAG and antitubulin antibodies. (D) Analysis of GCN4 expression by a β-galactosidase assay. Transformants of the Δfun12 strains carrying p180, together with the control (bars 1 and 2), eIF5B-WT (bars 3 and 4), or eIF5B-5A (bars 5 and 6) vector, were grown at 30°C in the absence (−3AT) (bars 1, 3, and 5) or presence (+3AT) (bars 2, 4, and 6) of 10 mM 3AT and assayed for β-galactosidase activity, as described in Materials and Methods. The graphs show averages and standard errors of six values in duplicate from three independent experiments. *, P < 0.001 by a t test. (E) Analysis of leaky scanning of the uORF1 start codon. Transformants of the Δfun12 strains carrying pM226, together with either the control (bar 1), eIF5B-WT (bar 2), or eIF5B-5A (bar 3) vector, were grown at 30°C and assayed for β-galactosidase activity. The data presented are the averages and standard errors of six values in duplicate from three independent experiments. *, P < 0.001 by a t test.
