Summary
Chemically induced pluripotent stem cells (CiPSCs) may provide an alternative and attractive source for stem cell-based therapy. Sufficient telomere lengths are critical for unlimited self-renewal and genomic stability of pluripotent stem cells. Dynamics and mechanisms of telomere reprogramming of CiPSCs remain elusive. We show that CiPSCs acquire telomere lengthening with increasing passages after clonal formation. Both telomerase activity and recombination-based mechanisms are involved in the telomere elongation. Telomere lengths strongly indicate the degree of reprogramming, pluripotency, and differentiation capacity of CiPSCs. Nevertheless, telomere damage and shortening occur at a late stage of lengthy induction, limiting CiPSC formation. We find that histone crotonylation induced by crotonic acid can activate two-cell genes, including Zscan4; maintain telomeres; and promote CiPSC generation. Crotonylation decreases the abundance of heterochromatic H3K9me3 and HP1α at subtelomeres and Zscan4 loci. Taken together, telomere rejuvenation links to reprogramming and pluripotency of CiPSCs. Crotonylation facilitates telomere maintenance and enhances chemically induced reprogramming to pluripotency.
Keywords: chemically induced pluripotent stem cells, telomeres, crotonic acid, Zscan4, genome stability
Graphical Abstract
Highlights
-
•
CiPSCs acquire telomere elongation after clonal formation with increasing passages
-
•
Both telomerase and recombination mechanisms are involved in the telomere elongation
-
•
Telomere damage and shortening can occur during late stage of lengthy induction
-
•
Crotonylation activates Zscan4 and promotes telomere elongation and CiPSC induction
In this article, Liu and colleagues show that telomere rejuvenation links to reprogramming and pluripotency of CiPSCs. Moreover, crotonylation induced by crotonic acid facilitates telomere maintenance and enhances chemically induced reprogramming to pluripotency.
Introduction
Somatic cells can be successfully induced by forced expression of four key transcription factors into induced pluripotent stem cells (iPSCs) that resemble embryonic stem cells (ESCs) (Takahashi and Yamanaka, 2006, Wernig et al., 2007, Zhao et al., 2009). The induction efficiency and quality of iPSCs can be significantly improved by various approaches, notably by small molecules (Huangfu et al., 2008a, Huangfu et al., 2008b, Li et al., 2013, Shi et al., 2008). In combination with small molecules or bone morphogenetic proteins, only Oct4 is needed for generation of iPSCs (Chen et al., 2011, Li et al., 2011, Zhu et al., 2010). High-content chemical screening has been attempted to achieve a goal of each transgenic factor being replaced with a small molecule that induces reprogramming (Ichida et al., 2009). Finally, somatic cells are successfully reprogrammed into pluripotent stem cells (PSCs) by pure small chemicals (Hou et al., 2013). The chemically induced PSCs (CiPSCs) presumably provide an alternative source with greater potential in stem cell-based therapy. Subsequently, two reports demonstrated successful generation of CiPSCs at higher efficiency by pure chemical compounds with slight modification of the recipe (Long et al., 2015, Zhao et al., 2015). Nevertheless, the reprogramming by complete chemicals is still at low efficiency and takes much longer than does OSKM-induced reprogramming (Maherali et al., 2007, Takahashi and Yamanaka, 2006).
Telomeres consist of repeated guanine-rich sequences and associated protein complexes known as shelterin that cap the end of chromosomes to maintain genomic stability (Blackburn, 2001, Palm and de Lange, 2008). Short telomeres can lead to cell senescence or tumorigenesis (Collado et al., 2007). Sufficiently long telomeres are important for unlimited self-renewal and pluripotency of PSCs (Huang et al., 2011, Marion et al., 2009). Short telomeres impair the differentiation capacity of ESCs (Pucci et al., 2013). Telomere length is maintained primarily by the ribonucleoprotein telomerase, a complex of a reverse transcriptase encoded by three core components: Tert (telomerase reverse transcriptase), template RNA Terc (essential RNA component), and dyskerin (Palm and de Lange, 2008). Telomerase is strongly expressed and required for telomere maintenance of mouse and human PSCs (Huang et al., 2011, Huang et al., 2014, Marion et al., 2009, Teichroeb et al., 2016, Wang et al., 2012). However, it remains elusive whether telomeres are appropriately reprogrammed and sufficiently elongated in CiPSCs.
We attempted to investigate telomere dynamics of CiPSCs generated based on the methods described recently (Long et al., 2015, Zhao et al., 2015). We found that CiPSCs acquire telomere lengthening with increasing passages. Surprisingly, telomeres suffer from erosion at late stages during extended periods of chemical induction, limiting reprogramming efficiency. We searched for compounds that can reduce telomere damage and shortening and thus improve chemical reprogramming. Promisingly, histone crotonylation induced by crotonic acid can alleviate telomere damage and shortening, enhancing the chemical induction efficiency.
Results
Generation of CiPSCs
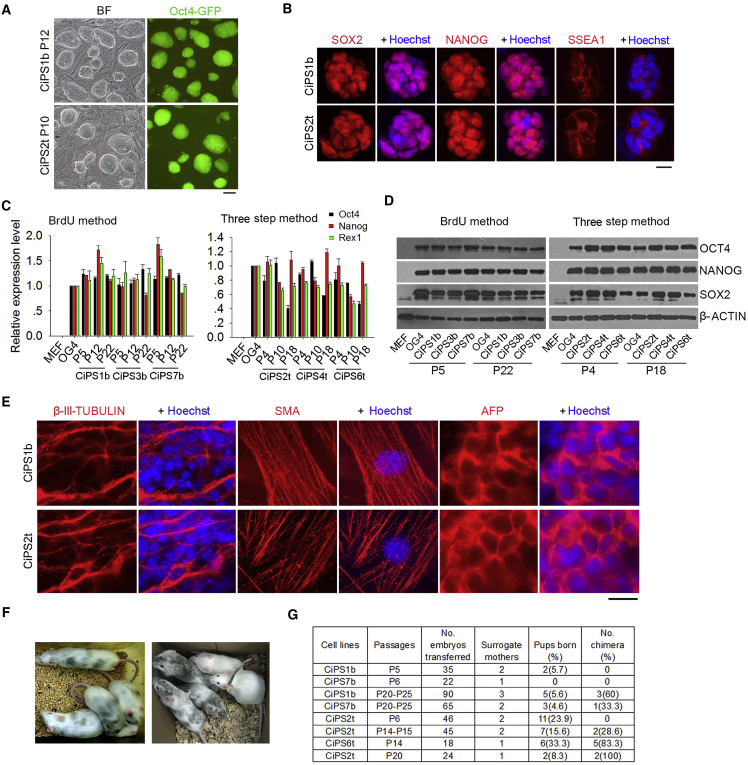
We attempted to generate CiPSCs following two recently published methods. One used a combination of seven small-molecule compounds (Hou et al., 2013), and bromodeoxyuridine (BrdU) (Long et al., 2015), herein referred to as the BrdU method. The other requires three stages to complete induction of CiPSCs, which undergo an extra-embryonic endoderm (XEN)-like state as an intermediate, and differs from the pathway of transcription factor-induced reprogramming, so is referred to as the three-step method (Zhao et al., 2015). Mouse embryonic fibroblasts (MEFs) were isolated from Oct4-GFP (OG2) transgenic mice harboring a GFP reporter driven by the distal Oct4 promoter and enhancer, activation of which indicates a naive state of pluripotency (Bao et al., 2009, De Los Angeles et al., 2015, Tang et al., 2010, Yeom et al., 1996). We successfully generated CiPSCs from OG2-MEFs following either the BrdU method (the randomly selected cell lines for further studies were CiPS1b, 3b, and 7b) or three-step method (cell lines named as CiPS2t, 4t, and 6t) (Figure S1A).
Continuous passages of ESC-like primary colonies established stable CiPSC lines that resembled typical ESC colonies in morphology, exhibiting large nuclei and nucleoli and clear compact clonal boundaries and expression of Oct4-GFP (Figures 1A and S1A), distinct from feeder fibroblasts. Colonies were stochastically picked and six established CiPSC lines chosen for further characterization of their pluripotency. By direct comparison with OG4 ESC lines established simultaneously from syngeneic background (Supplemental Experimental Procedures), CiPSCs exhibited pluripotency, as shown by expression at similarly high levels of key pluripotency factors OCT4, NANOG, SOX2, and Rex1, and surface marker SSEA1 by immunofluorescence microscopy, RT-qPCR, and western blot (Figures 1B, 1D, and S1C). Expression levels of pluripotency genes did not differ in CiPSCs at various passages (passage 4 [P4] to P22), like those of ESCs (OG4) at P4–5. CiPSCs also expressed high alkaline phosphatase activity (Figure S1D).
Figure 1.
Pluripotency and Differentiation Capacity of CiPSCs
(A) Representative morphology of CiPSCs generated by two methods (BrdU method, CiPSC1b at P12, and three-step method, CiPSC2t at P10) under bright field (BF) with phase contrast optics and expression of Oct4-GFP fluorescence. Scale bar represents 100 μm.
(B) Immunofluorescence microscopy of pluripotent markers SOX2, NANOG, and SSEA1. Scale bar represents 10 μm.
(C) mRNA expression levels by qPCR of Oct4, Nanog, and Rex1 in CiPSCs at various passages, compared with isogenic ESCs (OG4) and progenitor MEFs. Data represent mean ± SEM from three independent experiments.
(D) Protein levels of OCT4, NANOG, and SOX2 by western blot analysis of CiPSCs at earlier and advanced passages.
(E) Differentiation capacity in vitro of CiPSCs by immunofluorescence microscopy of three germ layer markers. Scale bar represents 10 μm.
(F) Left photo represents chimeras generated from the BrdU method and the right from the three-step method.
(G) Summary table showing percentage of chimeras generated from CiPSCs at different passages compared with OG4 ESCs. Chimeras (black and albino coat) were initially identified by coat color and some confirmed by microsatellite genotyping. CiPSC1b and 7b were generated using the BrdU method and CiPSC2t and the 6t by three-step method.
See also Figure S1.
These CiPSCs were able to differentiate into three embryonic germ layers by embryoid body formation in vitro, namely ectoderm, mesoderm, and endoderm (Figure 1E). We also tested the developmental pluripotency in vivo by injecting the CiPSCs into four- to eight-cell recipient albino embryos followed by embryo transfer. CiPSC1b and 7b cell lines and CiPSC2t and 6t cell lines at advanced passages efficiently generated chimeras by coat color (Figures 1F and 1G), but chimeras from these CiPSCs lines (n = 4 for BrdU method, and n = 10 for three-step method) failed to produce germline transmission following breeding with albino ICR mice for more than two rounds. Nevertheless, CiPSCs at earlier passages (P4 or P5) failed to form chimeras (Figure 1G).
These results validate that the CiPSCs do exhibit pluripotency in vivo and differentiation capacity in vitro. However, acquisition of developmental pluripotency of CiPSCs depends on the extended passages.
Telomeres of CiPSCs Rejuvenate with Increasing Passages
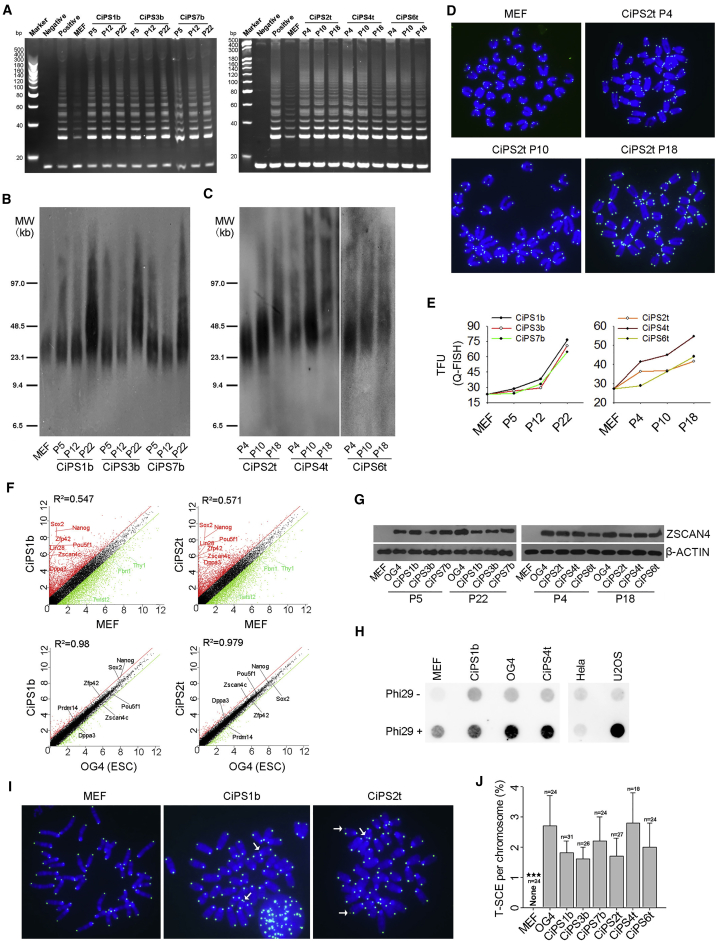
To investigate whether telomeres are elongated and associated with pluripotency of CiPSCs with increasing passages, three CiPSC lines generated by either method were analyzed for their telomere lengths at various passages, P4∼P6 (arbitrarily considered as early passages), P10∼P12 (mid passages), and P18∼P22 (late or advanced passages). Key telomerase genes Terc and Tert were expressed at higher levels in all CiPSC lines than in MEFs, and comparable with those of ESCs regardless of passages (Figure S2A). Higher expression levels of telomerase genes corroborated with higher telomerase activity in CiPSCs compared with MEFs (Figure 2A). TRF1, Pot1a/b, Rap1, and Tin2 components of the shelterin complex to safeguard robust telomeres (de Lange, 2005) also were upregulated in the CiPSCs (Figures S2B and S2C).
Figure 2.
Telomere Rejuvenates in CiPSCs with Passages
(A) Telomerase activity by telomeric repeat amplification protocol (TRAP) assay of CiPSCs during passages compared with progenitor cell MEFs. Lysis buffer served as negative control and OG4 ESCs as positive control.
(B and C) Telomere length distribution shown as TRF by Southern blot analysis of MEFs and CiPSCs following passages.
(D) Representative images displaying telomere FISH of MEF and CiPSCs at various passages. Blue, chromosomes stained with DAPI; green dots, telomeres.
(E) Line progression plot showing dynamics of relative telomere length by Q-FISH shown as telomere fluorescence unit (TFU) of MEFs and CiPSCs for each cell line from early to late passage. Left, BrdU method; right, three-step method.
(F) Scatterplots showing comparison of genome-wide transcription profile between CiPS1b or CiPS2t and ESC (OG4) or MEF. Parallel diagonal lines indicate 2-fold threshold in expression difference. Red, upregulated genes; green, downregulated genes.
(G) ZSCAN4 protein levels by western blot analysis. MEF and ESCs (OG4) served as negative and positive controls, respectively. β-ACTIN served as loading control.
(H) C-circle assay. Hela and U2OS served as negative and positive controls, respectively.
(I) Representative micrographs showing telomere sister chromatid exchange (T-SCE, white arrows) by chromosome orientation FISH analysis.
(J) Frequency of T-SCE. n, number of spread counted. ∗∗∗p < 0.001, compared with MEFs. Two independent experiments.
See also Figures S2 and S3.
Moreover, telomeres lengthened gradually in the formed CiPSCs and elongated further with increasing passages, estimated by telomere quantitative fluorescent in situ hybridization (Q-FISH) and also validated by telomere restriction fragment (TRF) analysis (Figures 2B and S2D). Interestingly, CiPSCs generated by the BrdU method initially exhibited only minor telomere elongation from P5 to P12, comparable with that of MEFs, but achieved robust telomere elongation by continuous passage and culture to P22 (Figures 2B, 2E, and S2D). CiPSCs obtained by the three-step method showed longer telomeres at early passage (e.g., P4), relative to those of MEFs, and gradually elongated telomeres from P4 to P18 (Figure 2C). Telomeres were modestly extended in most CiPSCs at earlier passage CiPSCs (CiPSC1b, 3b, 7b at P5 and CiPSC2t and 6t at P4) in comparison with those of the precursor MEFs, except for one cell line, CiPS4t, that possessed moderately longer telomere at P4 (Figures 2C, 2E, and S2D). Telomere lengths and the trend in their elongation estimated by the Q-FISH method corresponded with those measured by TRF.
These data suggest that telomeres of somatic cells are rejuvenated in the formed CiPSCs following passage and culture in vitro. Achievement of developmental pluripotency of CiPSCs at advanced passages is closely associated with sufficiently elongated telomeres and this also can be suggestive of full reprogramming.
CiPSCs Express Two-Cell Genes, Including Zscan4
By RNA-sequencing (RNA-seq) analysis, genome-wide gene expression profile differed remarkably between CiPSCs and MEFs but was similar between CiPSCs and ESCs (Figure 2F). CiPSCs highly expressed naive state marker genes, such as Zfp42/Rex1, Esrrb, Dppa3, and Prdm14, like ESCs (Nichols and Smith, 2009, Valamehr et al., 2014) (Figure 2F). Furthermore, CiPSCs expressed Zscan4 at higher levels but Twist2, Thy1, and Fbn1 genes for lineage differentiation at lower levels, in contrast to MEFs (Figure 2F). Zscan4 and other 2C-genes, including MuERVL, Tcstv1, and Tcstv3, were expressed at similarly high levels in CiPSCs and ESCs in contrast to MEFs (Figures 2F, 2G, and S2E). Proportion of Zscan4+ cells was comparable in CiPSCs and ESCs (P15) by both immunofluorescence microscopy and flow cytometry (Figures S2F–S2H). Zscan4 and MuERVL are expressed specifically in two-cell (2C) embryos and transiently in sporadic ESCs (1%–5%) at any given time, marking a transient 2C state of mouse ESCs (Macfarlan et al., 2012, Zalzman et al., 2010). Telomeres lengthen rapidly in 2C embryos and ZSCAN4 elongates telomeres promptly by recombination-based mechanisms and maintains genomic stability (Liu et al., 2007, Zalzman et al., 2010).
The single-stranded telomeric (CCCTAA)n DNA circles (C-circles) assay detects recombination-based alternative lengthening of telomeres (ALT) (Henson et al., 2009, Henson et al., 2017). We showed that C-circle levels were high in CiPSCs and ESCs, comparable with U2OS serving as ALT control, but low or minimal in Hela as negative controls, and in MEFs (Figure 2H). In addition, telomere sister chromatid exchange (T-SCE) has been implicated as telomere recombination-based ALT (Bailey et al., 2004, Bechter et al., 2004, Liu et al., 2007, Londono-Vallejo et al., 2004, Zalzman et al., 2010). Like iPSCs generated by the conventional induction method (Wang et al., 2012), CiPSC lines also displayed increased frequency of T-SCE at a level similar to that of ESCs, in contrast to MEFs (Figures 2I and 2J). These data suggest that the telomere recombination-based ALT mechanism is involved in telomere elongation of CiPSCs.
Although telomerase is activated earlier during induction, it is unclear whether levels of telomerase activity indeed influence induction of CiPSCs. To understand this, we attempted to generate CiPSCs using a three-step method from telomerase Terc-deficient (knockout, Terc−/−) MEFs, compared with heterozygous (HT) Terc+/− and wild-type (WT) MEFs. CiPSC primary colonies were obtained, and the efficiency of induction did not differ among Terc−/−, Terc+/−, and WT MEFs, based on alkaline phosphatase activity assay (Figures S3A and S3B). These data suggest that telomerase is dispensable for induction of CiPSCs. Despite lack or haploinsufficiency of telomerase activity, these CiPSCs still expressed pluripotency genes (e.g., Oct4, Sox2, Nanog, Zscan4, and SSEA1) at high levels, as did WT CiPSCs (Figures S3C–S3G). However, the telomerase-deficient CiPSCs showed telomere shortening, telomere loss, and chromosomal fusions, compared with WT CiPSCs generated simultaneously (Figures S3H and S3I), indicating that telomerase is essential for telomere maintenance of CiPSCs. This is also consistent with human iPSC induction, where telomerase gene mutation causes telomere shortening during induction, but activation of telomerase TERC by OCT4 acquires telomere rejuvenation and maintenance during passages after iPSC formation (Agarwal et al., 2010). Collectively, telomeres rejuvenate in CiPSCs following culture in vitro by passaging. Both telomerase and telomere recombination-based mechanism are required for telomere elongation in CiPSCs, like ESCs.
CiPSCs with Longer Telomeres Exhibit Higher Differentiation Potential
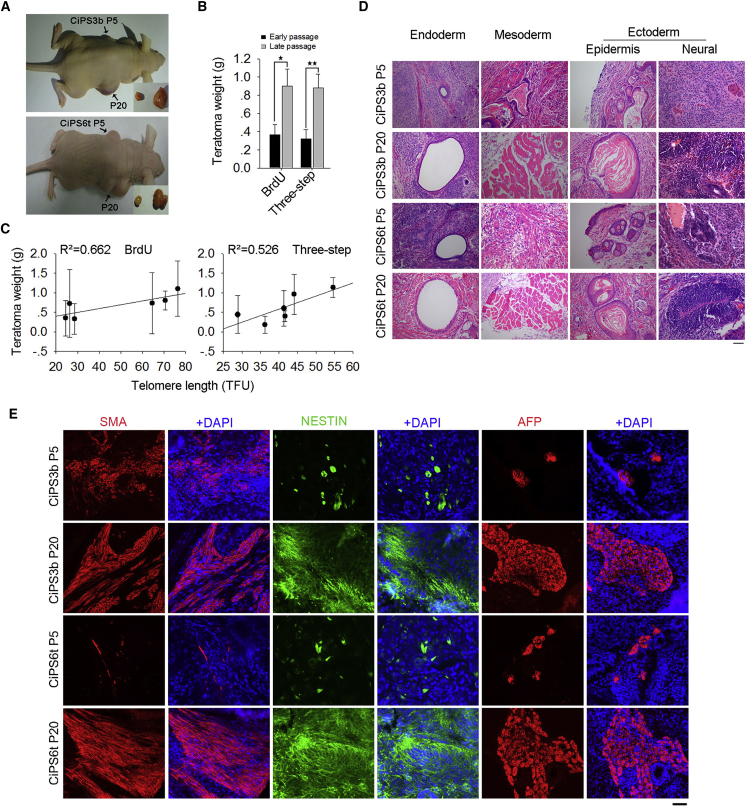
Sufficiently elongated telomeres may indicate the achievement of full reprogramming and differentiation capacity of CiPSCs. To test whether telomere lengths indicate differentiation capacity of CiPSCs in vivo, we performed a teratoma formation test by injection of CiPSCs at early and advanced passages under the skin of immunodeficient mice (Maherali and Hochedlinger, 2008). CiPSCs derived by either the BrdU or three-step method had comparatively short telomeres at early passages, slightly longer than those of progenitor cells, and achieved longer telomeres at relatively late passages (Figures 2B–2E). Remarkably, teratomas derived from CiPSCs at late passages with longer telomeres were larger and heavier than those of CiPSCs at early passages with shorter telomeres (Figures 3A and 3B). Linear regression analysis showed high correlation between telomere length and teratoma weight of CiPSCs (R2 = 0.662 for BrdU method and R2 = 0.526 for three-step method) (Figure 3C).
Figure 3.
Telomere Length Indicates Differentiation Capacity by Teratoma Formation
(A) Differentiation in vivo of CiPSCs at early and late passages by teratoma formation test.
(B) Weight of teratomas formed from CiPSCs, Mean ± SEM (n = 6 mice). ∗p < 0.05, ∗∗p < 0.01.
(C) Linear regression analysis showing high correlation between telomere length (TFU by Q-FISH) and teratomas weight of CiPSCs.
(D) Histology by H&E staining of teratoma tissues derived from early and late passage CiPSCs. Scale bar represents 50 μm.
(E) Immunofluorescence of the teratomas showing markers representative of three germ layers: NESTIN (ectoderm), SMA (mesoderm), and AFP (endoderm). Scale bar represents 100 μm.
Moreover, although the representative three embryonic germ layers, including epidermis and neural ectoderm, muscle (mesoderm), and gland epithelium (endoderm), were found in the teratomas formed from CiPSCs, the structures of the characteristic germ layers generally were smaller from CiPSCs at early passages than those of CiPSCs at late passages (Figure 3D). Immunofluorescence microscopy further validated that these teratomas expressed molecular markers representative of three germ layers, including NESTIN (ectoderm), smooth muscle actin (SMA) (mesoderm), and alfa-fetoprotein (AFP) (endoderm). However, these germ layers developed more extensively in teratomas formed from CiPSCs at late passages than did those at early passages (Figure 3E). Hence, by both histology and immunofluorescence microscopy, CiPSCs that acquire telomere lengthening and longer telomeres show higher differentiation potential in vivo.
Telomere Shortening during Late Stage of Lengthy Induction by Small Molecules
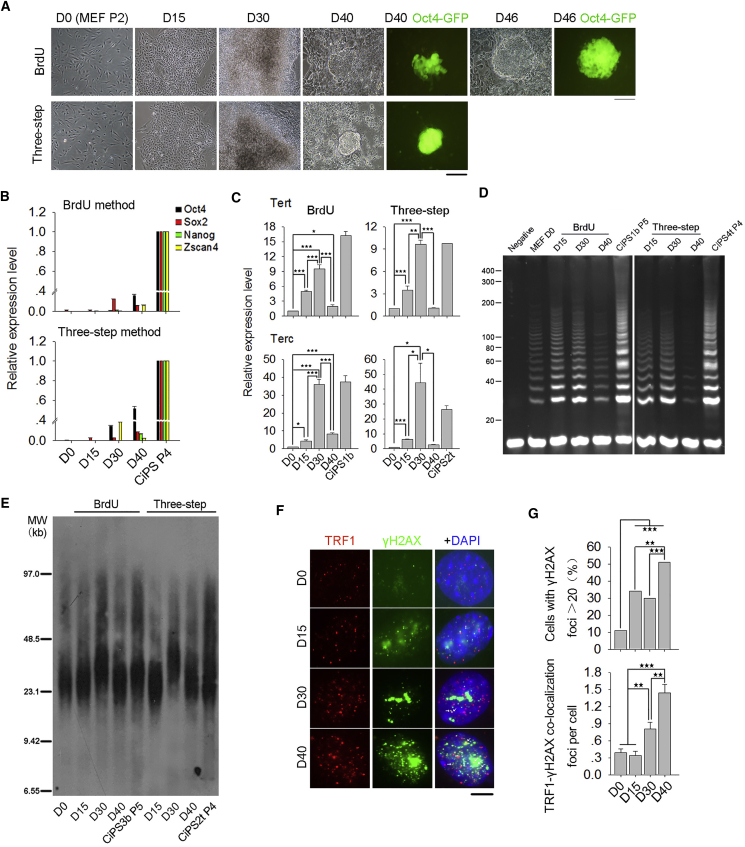
To understand telomere changes during long period of chemical induction, we collected cell samples on days 0 (MEFs), 15, 30, and 40 and compared them with CiPSCs at early passage, which served as positive control, and analyzed expression of genes important for pluripotency and telomerase, telomerase activity, and telomere lengths. Formation of the primary CiPSC colonies can be obtained 40 days after induction by complete small molecules, consistent with previous reports (Long et al., 2015, Zhao et al., 2015). The MEFs emerged as aggregates and achieved a XEN-like state by expression of marker genes Gata4, Gata6, and Sall4 15 days following induction (Figures 4A and S4A), in agreement with the findings that the chemical reprogramming process requires the early formation of XEN-like cells (Zhao et al., 2015). By the BrdU method, mRNA levels of Oct4, Sox2, and Zscan4 slightly increased by days 30 and 40, and Nanog was not readily detectable. By the three-step method, Oct4, Sox2, Nanog, and Zscan4 were expressed at variable levels but much lower than those of the formed CiPSCs (Figure 4B). Western blot confirmed that protein levels of OCT4, NANOG, SOX2, and ZSCAN4 were minimal or undetectable during induction, in contrast to the formed CiPSCs after primary clonal selections (Figure S4B). Notably, expression levels of telomerase genes Tert and Terc were already elevated by day 15 of induction and continued to increase to day 30, like those of the formed CiPSCs (Figure 4C). Thus telomerase genes are activated earlier than the pluripotent genes Oct4, Sox2, and Nanog (Figures 4B and 4C). The dynamics are consistent with those of conventional iPSC induction despite much less time required by OSKM-induced reprogramming (Wang et al., 2012).
Figure 4.
Telomere Dynamics during CiPSC Induction
(A) Morphological changes of MEFs during chemical induction under bright field with phase contrast optics or Oct4-GFP fluorescence. Compact colonies were formed at day 40 of induction by three-step method, earlier than with the BrdU method. Scale bar represents 100 μm.
(B) mRNA expression levels of Oct4, Sox2, Nanog, and Zscan4 during chemical induction.
(C) Relative expression levels of Tert and Terc during chemical induction.
(D) Telomerase activity by TRAP assay of MEFs and reprogramming cells during induction, compared with CiPSCs at P4 or P5. Lysis buffer as negative control.
(E) Telomere length distribution shown as TRF by Southern blot analysis during chemical induction (day 0–40) compared with CiPSCs. MW, molecular weight.
(F) Immunofluorescence showing co-staining of γH2AX (green) at telomeres (TRF1, red). Yellow (white arrows) in merged images indicates co-localized foci of TRF1 and γH2AX. Scale bar represents 5 μm.
(G) Quantification of TIFs. n = 100 cells counted.
Data represent mean ± SEM from three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
See also Figure S4.
However, expression levels of both Tert and Terc declined by day 40 of induction (Figure 4C). Correspondingly, telomerase activity was increased at day 15 and high at day 30 but decreased by day 40 (Figure 4D). In addition, TRF1 protein expression was gradually increased but also slightly reduced by day 40. TRF1 expression is increased during iPSC induction by the conventional method (Schneider et al., 2013). Moreover, levels of histone H3K9ac, H3K9me3, and H3K27me3 were increased at early stage of chemical induction, and H3K9ac and H3K27me3 were reduced at day 40 (Figure S4B).
Telomeres maintained and elongated during the early to mid stage of chemical induction (days 0–30) (Figure 4E) coincided with increased telomerase activity (Figure 4D). Unexpectedly, telomeres shortened during the late stage of CiPSC induction (days 30–40), when the telomerase activity also was decreased (Figure 4D). These outcomes prompted us to investigate whether reprogramming cells undergo telomere damage after prolonged induction by examining telomere dysfunction-induced damage foci (TIFs) indicated by co-localized foci of phosphorylated H2AX with TRF1 at telomeres (Dan et al., 2014, Takai et al., 2003). Indeed, co-localized foci of γH2AX and TRF1 were increased by day 40, and the proportion of the cells exhibiting more than 20 γH2AX foci also was elevated (Figures 4F and 4G). Thus, DNA and telomere damage may occur during the late stage of extended induction.
Few apoptotic cells were found on day 0 and day 15, but number of apoptotic cells with fragmented nuclei increased from day 30 to day 40 (Figures S4C and S4D). These data suggest that reduced telomerase, increased telomere damage, and apoptosis together may be responsible for the telomere erosion during prolonged periods of chemical induction, and this can limit CiPSC formation.
Histone Crotonylation Activates Zscan4 and Maintains Telomeres during Chemical Induction
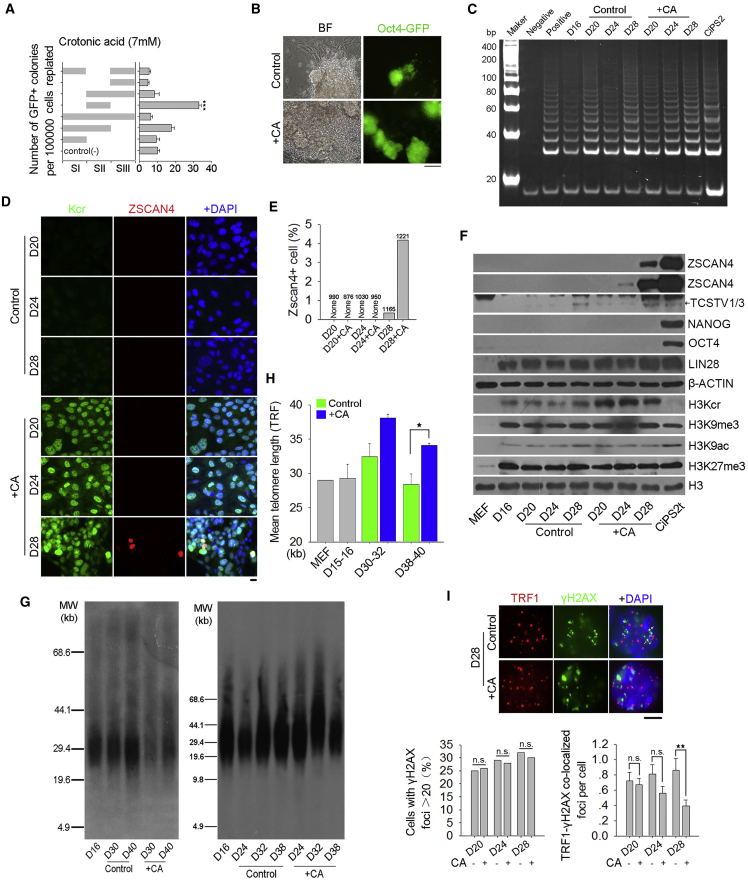
Zscan4 is not effectively activated during chemical induction until in the formed CiPSCs following passages (Figures 4B and S4B), like conventional iPSC induction (Wang et al., 2012). Forced expression of Zscan4, in combination with Yamanaka factors, greatly reduces DNA damage response, and increases telomere elongation and quality of iPSCs (Jiang et al., 2013). Heterochromatic histone assembly, such as HP1 and H3K9me3 at telomeres/sub-telomeres, can regulate Zscan4 expression (reviewed by Liu, 2017). We searched for a variety of histone modification by small molecules, including trichostatin A, valproic acid, and sodium butyrate, that can activate Zscan4 expression in mESCs (Dan et al., 2014, Dan et al., 2015), but none of them can effectively activate Zscan4 during chemical induction (data not shown). Excitingly, we found that a chemical compound, crotonic acid (Figure S5A), which mediates lysine crotonylation (Kcr) (Tan et al., 2011, Wei et al., 2017a, Wei et al., 2017b), can induce protein epigenetic modification and force Zscan4 expression. Initially, mESCs were treated with 5 mM or 10 mM crotonic acid (Figure S5B). 2C genes, including Zscan4, Tcstv1, and Tcstv3, were rapidly upregulated following two passages (Figures S5C and S5D), while genes for pluripotency (Oct4, Sox2, and Nanog) and telomerase Tert were downregulated (Figures S5C and S5D). Proportion of Zscan4+ cells also was increased (Figure S5E). Moreover, level of histone lysine crotonylation (Kcr), rather than other histone modifications (e.g., H3K27me3, H3K9me3, or H3K9ac), was elevated (Figures S5D and S5F).
Further, we tested whether crotonic acid could also activate Zscan4 during chemical induction (Figure 5A). Initially, MEFs were treated with crotonic acid or vehicle controls for the whole period of induction using the three-step method because of its higher induction efficiency (Figure S1B), and were examined for colonies positive for Oct4-GFP fluorescence by the end of induction. Number of Oct4-GFP-positive colonies was not affected by crotonic acid. Then, we treated MEFs with crotonic acid at various periods, illustrated in Figure 5A, and observed that only the treatment for the period of stage II (days 16–28) effectively promoted Oct4-GFP colony formation. The Oct4-GFP colonies increased 3-fold in crotonic acid-treated-cells compared with non-treated cells (Figures 5A, 5B, S6A, and S6B). Expression of Oct4, Nanog, Sox2, Lin28, and Terc and Tert was not affected by crotonic acid during induction (Figures 5F and S6C), consistent with telomerase activity (Figure 5C). Remarkably, addition of crotonic acid from day 16 resulted in ZSCAN4 activation by day 28 (Figures 5D–5F and S6D). Histone lysine crotonylation (Kcr) was increased by crotonic acid treatment, while H3K9ac, H3K27me3, and H3K9me3 levels were not changed (Figures 5F and S6D). 2C genes TCSTV1/3, which can elongate telomeres in mESCs (Zhang et al., 2016), also were upregulated after crotonic acid treatment (Figure 5F).
Figure 5.
Crotonic Acid Addition at Stage II Activates Zscan4, Maintains Telomeres, and Promotes CiPSC Generation
(A) Durations of crotonic acid (7 mM) addition at different time points during chemical induction and assessment of Oct4-GFP-positive colonies on day 40. Left panel shows different time points during reprogramming. Stage I (SI) represents days 0–15; stage II (SII) represents days 16–28; stage III (SIII) represents days 29–40. Right panel shows number of Oct4-GFP-positive colonies on day 40; 100,000 cells were re-plated on day 12 following induction. ∗∗∗p < 0.001, compared with control.
(B) Morphology by phase contrast optics and Oct4-GFP fluorescence of primary CiPSC colonies on day 40 induced with or without crotonic acid (CA) at stage II. Scale bar represents 100 μm.
(C) Telomerase activity by TRAP assay. Lysis buffer and ESCs served as negative and positive controls, respectively.
(D) Immunofluorescence microscopy of ZSCAN4 and Kcr (lysine crotonylation) expression. Scale bar represents 10 μm.
(E) Percentage of Zscan4-positive cells. Number of cells counted is shown above the bar.
(F) Western blot analysis of protein levels. MEF and CiPSCs served as controls.
(G) Telomere length distribution shown as TRF by Southern blot analysis.
(H) Quantification of telomere length by TRF in three repeats using TeloTool (Gohring et al., 2014). ∗p < 0.05.
(I) Immunofluorescence of γH2AX (green) at telomeres (TRF1, red). Yellow foci in the merged images indicate TRF1-γH2AX co-localization. Scale bar represents 5 μm. Bottom panel, quantification of TIFs. n = 100 cells counted. ∗∗p < 0.01, n.s., not significant.
Data represent mean ± SEM from three independent experiments. See also Figure S5.
Consistently, addition of crotonic acid at stage II rapidly lengthened telomeres following Zscan4 upregulation, compared with the control, and maintained telomeres at the late stage of induction (Figures 5G and 5H). In addition, number of γH2AX-positive cells did not differ at stage II between crotonic acid-treated and non-treated cells. However, co-localization foci (TIFs) of γH2AX and TRF1 were much reduced on day 28 by crotonic acid (Figure 5I).
These data suggest that addition of crotonic acid at stage II (days 16–28) promotes CiPSC generation. Crotonic acid activates Zscan4, reduces telomeric damage, and maintains telomere length during chemically induced reprogramming.
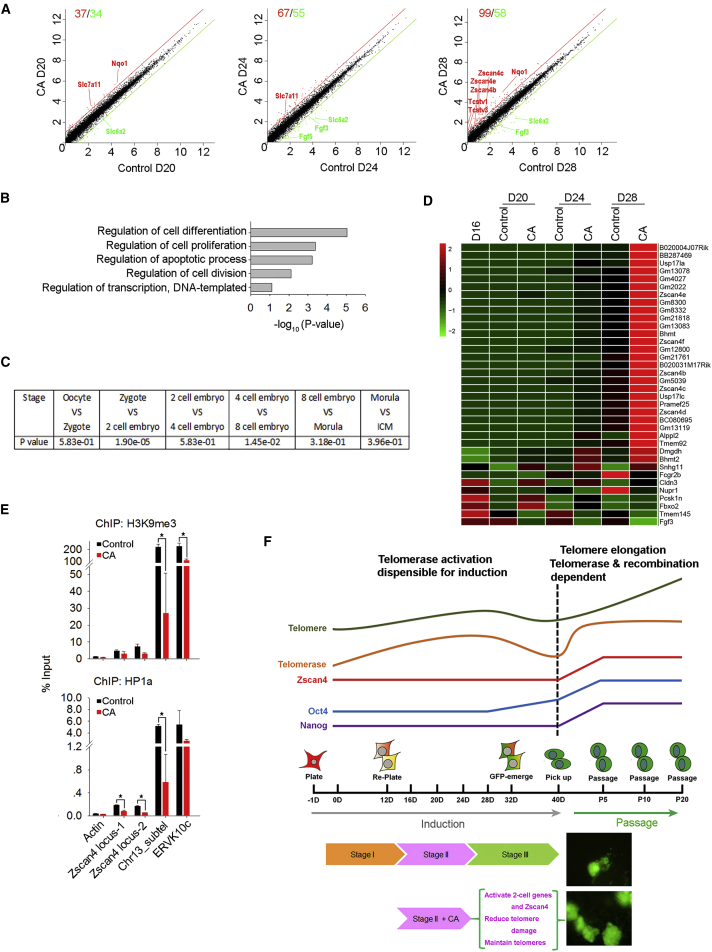
Crotonic Acid Activates 2C Genes and Increases T-SCE during Chemical Induction
To better understand the mechanisms underlying crotonic acid in facilitating chemically induced reprogramming, we performed RNA-seq analysis of cell samples by days 20, 24, and 28 without or with crotonic acid treatment. More genes were upregulated than downregulated in crotonic acid-treated cells compared with controls (Figure 6A). Numbers of differentially expressed genes gradually increased from day 20 to day 28 following induction (Figure 6A). These genes were enriched in regulation of cell differentiation, cell proliferation, and apoptosis (Figure 6B). RNA-seq data also verified that Zscan4 was prominently upregulated at day 28 following addition of crotonic acid on day 16 (Figures 6A and 5D–5F).
Figure 6.
RNA-Seq Reveals Crotonic Acid-Induced Activation of 2C Genes
(A) Scatterplots showing global differential gene expression profile (fold ≥ 2.0) of reprogramming cells on days 20–28 following treatment with CA in comparison with controls. Red, upregulated genes; green, downregulated genes in CA-treated cells. Average from two independent experiments.
(B) Gene ontology analysis of differentially expressed genes in cells treated with crotonic acid, compared with controls.
(C) Fisher's exact test between relatively upregulated differential genes at early embryo developmental stages from single-cell RNA-seq data (Fan et al., 2015) and all 196 differential genes from our RNA-seq data (p adjusted < 0.01).
(D) Heatmap highlighting highly expressed 2C genes found in CA-treated cells compared with controls.
(E) ChIP-qPCR analysis of H3K9me3 and HP1α abundance at Zscan4, Chr13_subtel (sub-telomere region of no. 13 chromosome), and ERVK10c loci following treatment with crotonic acid for 12 days during stage II of induction. β-actin served as negative control. Data represent mean ± SD from three independent experiments. ∗p < 0.05.
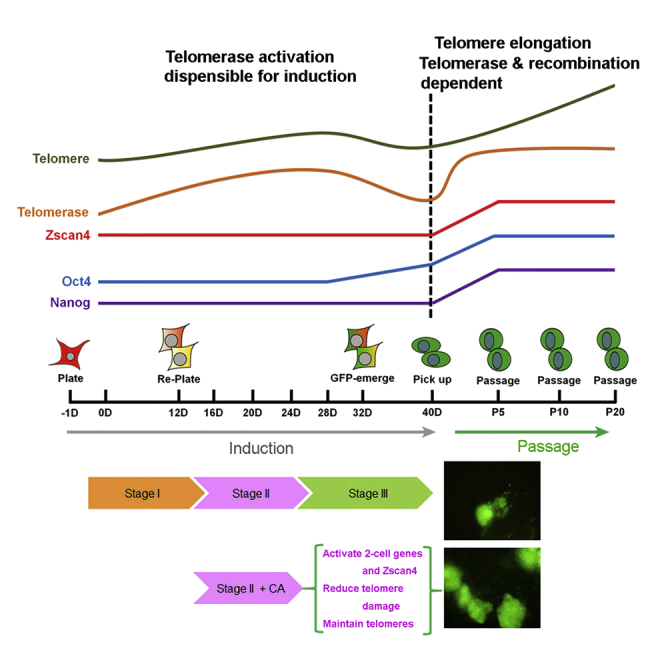
(F) Schematic diagram illustrating the process of CiPSC induction following addition of crotonic acid, which activates 2C genes including Zscan4, reduces telomere and DNA damage, and maintains telomeres, promoting CiPSC formation.
See also Figure S6.
We compared all 196 differentially expressed genes with single-cell RNA-seq data of mouse preimplantation embryos (Fan et al., 2015). Interestingly, by clustering of the differentially expressed genes to those expressed at cleavage stage of early embryo development, these genes were significantly enriched in the 2C (2C) embryo (p = 1.90 × 10−5), compared with other early embryonic development stages from oocyte to inner cell mass (ICM) of blastocysts (Figure 6C). Most of the 36 2C genes overlapped with the single-cell RNA-seq data were upregulated during induction by day 24 and noticeably by day 28 (Figure 6D). Additional comparison of our RNA-seq data with those of 2C genes (Macfarlan et al., 2012) substantiated that 2C genes were gradually upregulated following addition of crotonic acid during stage II of chemical induction (Figure S6E). The differentially expressed 2C genes and their gradual activation were further confirmed by qPCR (Figure S6F). However, crotonic acid reduced cell proliferation during the stage II reprogramming period (Figure S6G). To test whether crotonylation regulates local heterochromatic distribution at 2C gene regions, we performed a chromatin immunoprecipitation (ChIP)-qPCR experiment using H3K9me3 and HP1α antibodies and the primers specific for the promoter-proximal regions of Zscan4 loci, sub-telomere of chromosome 13, and the ERVK10c locus (Dan et al., 2014, Dan et al., 2015). Crotonylation decreased levels of H3K9me3 and HP1α at these loci following treatment with crotonic acid for 12 days (Figure 6E).
Moreover, crotonic acid increased the frequency of T-SCE at a level similar to that of CiPSCs, compared with untreated reprogramming cells at day 28 (Figures S6H and S6I). Addition of crotonic acid in CiPSCs further increased the incidence of T-SCE (Figures S6H and S6I). Increased ZSCAN4 expression levels are associated with higher frequency of T-SCE.
These data show that crotonic acid activates a series of 2C genes in the middle of chemical induction, and, together with crotonylation-mediated Zscan4 activation, increases T-SCE and telomere maintenance, and promotes CiPSC generation (Figure 6F).
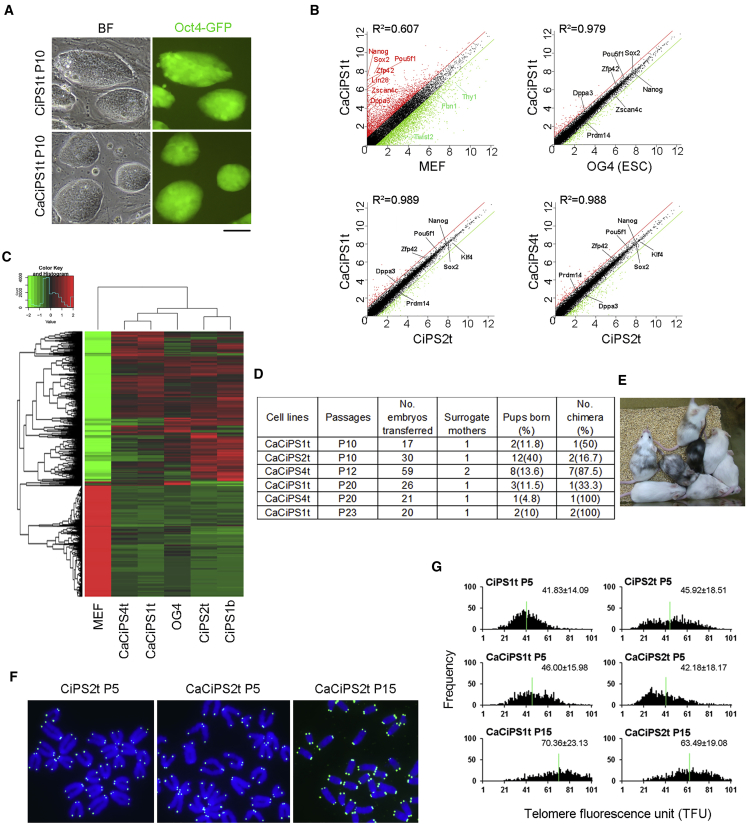
Crotonic Acid-Facilitated CiPSCs Possess High Pluripotency
The question was whether CiPSCs generated by addition of crotonic acid exhibit normal pluripotency. We characterized the CiPSCs derived by addition of crotonic acid (CaCiPSCs). Continuous passaging of the CaCiPSC primary colonies achieved stable CiPSC lines with Oct4-GFP fluorescence resembling typical ESC colonies in morphology, with large nuclei and nucleoli and clear, compact clonal boundaries (Figure 7A). Importantly, the majority of CaCiPSCs maintained normal karyotypes (Figure S7A). Moreover, CaCiPSCs expressed multiple pluripotent markers (Oct4, Nanog, Sox2, Lin28, Zscan4, and SSEA1) and also the naive state markers Stella, Prdm14, Rex1, and Tbx3 (Nichols and Smith, 2009, Valamehr et al., 2014), comparable with those of ESCs (Figures S7B–S7D).
Figure 7.
Generation and Characterization of CaCiPSCs Induced by Addition of Crotonic Acid at Stage II
(A) Morphology under bright field with phase contrast optics and Oct4-GFP fluorescence of CiPSCs generated without CA and CaCiPSCs with CA. Scale bar represents 100 μm.
(B) Scatterplots comparing transcriptome profile of CaCiPS1t from CA-treated cells, CiPS2t without CA treatment, OG4 ESCs, and MEFs. Parallel diagonal lines indicate 2-fold threshold.
(C) Hierarchical clustering of transcriptome profile of MEFs, OG4 ESCs, CiPS1b, CiPS2t, CaCiPS1t, and CaCiPS4t.
(D and E) Summary table showing efficiency of chimera generation from CaCiPSCs at mid and late passages following injection into four- to eight-cell albino embryos (D). Chimeras were initially identified by coat color (E) and confirmed by microsatellite genotyping.
(F) Representative telomere FISH images. Blue, chromosomes stained with DAPI; green dots, telomeres.
(G) Histogram displaying distribution of relative telomere length shown as TFU by telomere Q-FISH. Medium telomere length (green lines) is shown as mean ± SD above each panel.
See also Figure S7.
RNA-seq analysis revealed that the transcriptome profile of CaCiPS1t resembled that of ESCs (OG4), with a correlation coefficient (R2) of 0.979 but was quite different from that of MEF (Figure 7B). Expression levels of pluripotency genes, naive marker genes, and Zscan4 did not differ between CaCiPSCs and ESCs. Expression profile of CaCiPS1t and CaCiPS4t also was very similar to that of CiPS2t, and expression levels of pluripotency genes and naive marker genes did not differ between CaCiPSCs and CiPSCs (Figure 7B). All CiPSCs clustered with ESCs but not with the progenitor MEF cells (Figure 7C). Moreover, two CaCiPS cell lines, CaCiPS4t and CaCiPS1t, preferentially clustered with ESC OG4, relative to CiPS2t and CiPS1b (Figure 7C).
Furthermore, we tested the developmental potency by injection of CaCiPSCs into four- or eight-cell embryos from Balb/c albino mice. Chimeras were obtained with high efficiency by coat color (Figures 7D and 7E). However, cell lines varied in their contribution to chimeras. For instance, CaCiPSC2t generated chimeras at a very low rate. Chimerism derived from CaCiPSC4t was high based on coat color and microsatellite genotyping (Figures 7E and S7F). Although CaCiPSCs can contribute to gonad in the chimeras (Figure S7F), they also failed to produce germline transmission following breeding of chimeras (n = 10) with albino ICR mice for more than two rounds.
We also measured telomere lengths of CaCiPSCs at early and advanced passages. We picked up primary CaCiPSC and CiPSC colonies derived at the same time from the same batch of OG2-MEFs and passaged them in vitro. Telomere lengths varied among the cell lines but generally were not elongated much at P5 compared with MEFs (Figure S7E), and this may indicate heterogeneity of primary CiPSC clones. By Q-FISH, telomere lengths also were indistinguishable among CiPS1t, CiPS2t, CaCiPS1t, and CaCiPS2t at P5 (Figures 7F and 7G). Nevertheless, telomeres lengthened significantly in both CaCiPS cell lines with increasing passages from P5 to P15 (CaCiPS1t, from 46.00 ± 15.98 to 70.36 ± 23.13 telomere fluorescence units [TFU]; CaCiPS2t, from 42.18 ± 18.17 to 63.49 ± 19.08 TFU) (Figure 7G). Hence, CaCiPSCs manifested high pluripotency, in association with telomere rejuvenation with passages.
To further explore the effect on genome stability of BrdU or crotonic acid addition during chemical reprogramming, we performed the whole-exome sequencing analysis. We compared BrdU-treated MEF cells with control MEF cells following culture for 12 days simultaneously and did not observe somatic point mutations in protein-coding regions (Tables S4 and S5). Moreover, we characterized the protein-coding region of six CiPSC lines by three induction methods that generated chimeras at single-nucleotide resolution. The number of point mutations of these CiPSCs was similar to what had been previously reported for iPSCs (Figure S7G) (Gore et al., 2011). The CiPS1b and CiPS7b (BrdU method) cell lines had slightly more point mutations in coding regions (13 and 29) than did CiPS2t and CiPS6t (three-step method, 12 and 9) and CaCiPS2t and CaCiPS4t (crotonic acid method, 17 and 9) lines (Figure S7G, Tables S4 and S5). These mutated genes, however, did not cluster in a specific functional pathway.
Discussion
We show that telomeres of CiPSCs elongate with passages after clonal formation. Telomere elongation with increasing passage depends on both telomerase and recombination-based ALT mechanisms (Figure 6F). Moreover, CiPSCs express 2C genes including Zscan4, Tcstv1/3, and MuERVL, like ESCs. Successful generation of CiPSCs further supports the notion that reprogramming and rejuvenation of telomeres are intimately linked to somatic reprogramming to induction of pluripotency. However, telomere damage occurs and telomeres shorten at late stage during prolonged chemical induction. Crotonylation induced by crotonic acid activates 2C genes, notably Zscan4, and increases T-SCE, which maintains telomeres and reduces telomere damage during chemical induction, improving induction efficiency (Figure 6F).
Currently, the low efficiency in achieving Oct4-GFP fluorescence colonies and particularly the lengthy induction still limit CiPSC generation. Oct4 expression levels are very low during induction and can only be seen at late stage by qPCR analysis. OCT4 protein levels are too low to be detected by western blot during induction compared with the formed CiPSCs. Importantly, Oct4 distal enhancer activation indicates achievement of sufficient reprogramming and is essential for achieving a naive pluripotency network (Bao et al., 2009, De Los Angeles et al., 2015, Gafni et al., 2013). Notably, telomerase activates earlier than do pluripotency genes Oct4 and Sox2. Early activation of telomerase might result from CHIR99021 used at high concentration (seven times more than routinely used), which activates Wnt/β-catenin. Wnt/β-catenin signaling in turn can directly activate telomerase (Hoffmeyer et al., 2012). Our data suggest that Oct4 could be expressed in only a few cells, revealed by the distal Oct4-GFP fluorescence during the late stage of chemical induction, such that the protein level in a cell population is minimal by western blot. Thus, timing to activate Oct4 becomes essential for further enhancing CiPSC induction.
Also, Nanog is not activated well during chemical induction until CiPSC clonal formation and passages (Figure 6F). Nanog mediates acquisition of both embryonic and induced pluripotency (Silva et al., 2009). Moreover, biallelic expression of Nanog is required in the timely maturation of the ICM into a fully reprogrammed pluripotent epiblast (Miyanari and Torres-Padilla, 2012). Inhibiting Tgf-β signaling promotes the completion of reprogramming through induction of Nanog (Ichida et al., 2009), and Nanog overexpression can accelerate reprogramming (Hanna et al., 2009). Presumably, chemicals that can activate Nanog earlier likely increase CiPSC induction.
Unexpectedly, telomeres shorten, coincident with telomere damage, and incidence of apoptosis also is increased during the late stage of lengthy induction by pure chemicals (Figure 6F). Fewer cells are likely to be selected and survive during reprogramming induction. This is in contrast to conventional iPSCs induced by Yamanaka factors, by which telomeres continue to elongate or at least maintain their length without shortening during the induction period (Wang et al., 2012). Thus, chemicals that can alleviate telomere damage and shortening during the late stage of induction would enhance CiPSC generation.
Activation of Zscan4 during induction indeed reduces telomere damage and improves iPSC induction and quality (Hirata et al., 2012, Jiang et al., 2013). We find that crotonylation facilitated by crotonic acid can activate 2C genes and increase T-SCE during induction and that increased expression of ZSCAN4 reduces telomere damage and improves telomere maintenance, increasing CiPSC induction efficiency (Figure 6F).
Our data suggest that crotonic acid activates Zscan4 and 2C genes, likely by reducing heterochromatic histones (e.g., H3K9me3 and HP1α) at telomeres/subtelomeres. This is consistent with the findings that histone crotonylation accumulates with HP1γ (Tan et al., 2011) and reduces HP1α at heterochromatin (Wei et al., 2017b), and reduction of H3K9me3 leads to activation of 2C genes at subtelomeres; e.g., Zscan4 and Tcstv1 (Dan et al., 2014, Dan et al., 2015). More experiments are needed to understand whether crotonylation directly or indirectly regulates the processes. Additionally, histone crotonylation has critical and broad function as histone acetylation in transcription (Wei et al., 2017a). Moreover, crotonylation improves CiPSC clone formation at stage II during middle induction, implying that XEN state is primed for action by crotonic acid. Continuous treatment with crotonic acid is not needed after Zscan4 activation. Likewise, forced expression of Zscan4 is required only for the first few days of conventional iPSC formation (Hirata et al., 2012).
Telomere maintenance during induction is one of the key elements for successful reprogramming to pluripotency. Histone crotonylation by crotonic acid is one example that protects telomeres by activating 2C genes and Zscan4 and increasing T-SCE-based ALT-like activity, as shown here. Human iPSCs and ESCs generated thus far have not been demonstrated to exhibit ALT-like mechanisms, unlike mouse ESCs/iPSCs. It is unclear whether this is related to the pluripotency status of ESCs/iPSCs, as presumably conventional human iPSCs/ESCs are more like mouse epiblast stem cells or primed states than naive mouse iPSCs/ESCs (Liu, 2017).
We anticipate that fine-tuning of treatment time and concentration of crotonic acid in combination with other chemicals that can facilitate early activation of Oct4 distal enhancer and promoter and of Nanog may accelerate the reprogramming and improve CiPSC induction.
Experimental Procedures
Mice and Cell Culture
Mice were housed and cared for in the College Animal Facility and the use of mice was approved by the Institutional Animal Care and Use Committee at Nankai University. All the animal experiments were performed following the ethical guidelines approved by the Tianjin Animal Management Committee. ESCs were cultured on mitomycin C-inactivated MEF feeder cells in the ESC medium, which was changed daily, and cells routinely passaged every 2 days. For culture of CiPSCs, 2i (3 μM CHIR99021 and 1 μM PD0325901) were added to the medium. The ESC culture medium consisted of knockout DMEM (Invitrogen) with 20% FBS (Hyclone), 1000 U/mL mouse leukemia inhibitory factor (LIF; ESG1107; Millipore), 0.1 mM non-essential amino acids, 0.1 mM β-mercaptoethanol, 1 mM L-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/mL).
CiPSC Induction from MEFs
MEFs were isolated from OG2 mice. Isolated OG-MEF cells at early passages (up to P3) were used for chemical induction following the method described (Zhao et al., 2015). The details are described in Supplemental Experimental Procedures.
Library Preparation and RNA Sequencing
A total amount of 3 μg of RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, United States) following manufacturer's recommendations and index codes were added to attribute sequences to each sample. More details are described in the Supplemental Experimental Procedures.
Differential Gene Expression Analysis
For DESeq with biological replicates, differential expression analysis of two groups (two biological replicates per condition) was performed using the DESeq R package (1.18.0). More details are described in the Supplemental Experimental Procedures.
ChIP-qPCR Analysis
ChIP-qPCR analysis was performed as described previously (Dan et al., 2014, Dan et al., 2015).
More methods are detailed in the Supplemental Experimental Procedures.
Author Contributions
H.F. conducted most experiments, analyzed the data, and prepared the manuscript. C.-l.T., X.Y., X.S., H.W., and Y.L. conducted part of the experiments or provided reagents. L.L. conceived the project, designed experiments, and wrote and revised the manuscript.
Acknowledgments
We thank Yuan Long and Xin Xie for kind assistance with chemical reprogramming experiments, Weiyu Zhang and Xinyi Lu for help with analysis of RNA-seq data, and Feng Wang and Jinghua Yuan for C-circle experiments and critical reading of the manuscript. This work was supported by the National Natural Science Foundation of China (31430052, 31571546) and PCSIRT (no. IRT13023).
Published: May 31, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and five tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.05.003.
Accession Numbers
The accession number for the RNA-seq data used in this study is GEO: GSE113921, and the number for whole-exome sequencing data is NCBI Sequence Read Archive: SRP144105.
Supplemental Information
References
- Agarwal S., Loh Y.H., McLoughlin E.M., Huang J., Park I.H., Miller J.D., Huo H., Okuka M., Dos Reis R.M., Loewer S. Telomere elongation in induced pluripotent stem cells from dyskeratosis congenita patients. Nature. 2010;464:292–296. doi: 10.1038/nature08792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S.M., Brenneman M.A., Goodwin E.H. Frequent recombination in telomeric DNA may extend the proliferative life of telomerase-negative cells. Nucleic Acids Res. 2004;32:3743–3751. doi: 10.1093/nar/gkh691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S., Tang F., Li X., Hayashi K., Gillich A., Lao K., Surani M.A. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature. 2009;461:1292–1295. doi: 10.1038/nature08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechter O.E., Zou Y., Walker W., Wright W.E., Shay J.W. Telomeric recombination in mismatch repair deficient human colon cancer cells after telomerase inhibition. Cancer Res. 2004;64:3444–3451. doi: 10.1158/0008-5472.CAN-04-0323. [DOI] [PubMed] [Google Scholar]
- Blackburn E.H. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- Chen J., Liu J., Yang J., Chen Y., Chen J., Ni S., Song H., Zeng L., Ding K., Pei D. BMPs functionally replace Klf4 and support efficient reprogramming of mouse fibroblasts by Oct4 alone. Cell Res. 2011;21:205–212. doi: 10.1038/cr.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M., Blasco M.A., Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Dan J., Liu Y., Liu N., Chiourea M., Okuka M., Wu T., Ye X., Mou C., Wang L., Wang L. Rif1 maintains telomere length homeostasis of ESCs by mediating heterochromatin silencing. Dev. Cell. 2014;29:7–19. doi: 10.1016/j.devcel.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J., Yang J., Liu Y., Xiao A., Liu L. Roles for histone acetylation in regulation of telomere elongation and two-cell state in mouse ES cells. J. Cell. Physiol. 2015;230:2337–2344. doi: 10.1002/jcp.24980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- De Los Angeles A., Ferrari F., Xi R., Fujiwara Y., Benvenisty N., Deng H., Hochedlinger K., Jaenisch R., Lee S., Leitch H.G. Hallmarks of pluripotency. Nature. 2015;525:469–478. doi: 10.1038/nature15515. [DOI] [PubMed] [Google Scholar]
- Fan X., Zhang X., Wu X., Guo H., Hu Y., Tang F., Huang Y. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 2015;16:148. doi: 10.1186/s13059-015-0706-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni O., Weinberger L., Mansour A.A., Manor Y.S., Chomsky E., Ben-Yosef D., Kalma Y., Viukov S., Maza I., Zviran A. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- Gore A., Li Z., Fung H.L., Young J.E., Agarwal S., Antosiewicz-Bourget J., Canto I., Giorgetti A., Israel M.A., Kiskinis E. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohring J., Fulcher N., Jacak J., Riha K. TeloTool: a new tool for telomere length measurement from terminal restriction fragment analysis with improved probe intensity correction. Nucleic Acids Res. 2014;42:e21. doi: 10.1093/nar/gkt1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J., Saha K., Pando B., van Zon J., Lengner C.J., Creyghton M.P., van Oudenaarden A., Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson J.D., Cao Y., Huschtscha L.I., Chang A.C., Au A.Y., Pickett H.A., Reddel R.R. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat. Biotechnol. 2009;27:1181–1185. doi: 10.1038/nbt.1587. [DOI] [PubMed] [Google Scholar]
- Henson J.D., Lau L.M., Koch S., Martin La Rotta N., Dagg R.A., Reddel R.R. The C-circle assay for alternative-lengthening-of-telomeres activity. Methods. 2017;114:74–84. doi: 10.1016/j.ymeth.2016.08.016. [DOI] [PubMed] [Google Scholar]
- Hirata T., Amano T., Nakatake Y., Amano M., Piao Y., Hoang H.G., Ko M.S. Zscan4 transiently reactivates early embryonic genes during the generation of induced pluripotent stem cells. Sci. Rep. 2012;2:208. doi: 10.1038/srep00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeyer K., Raggioli A., Rudloff S., Anton R., Hierholzer A., Del Valle I., Hein K., Vogt R., Kemler R. Wnt/beta-catenin signaling regulates telomerase in stem cells and cancer cells. Science. 2012;336:1549–1554. doi: 10.1126/science.1218370. [DOI] [PubMed] [Google Scholar]
- Hou P., Li Y., Zhang X., Liu C., Guan J., Li H., Zhao T., Ye J., Yang W., Liu K. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- Huang J., Wang F., Okuka M., Liu N., Ji G., Ye X., Zuo B., Li M., Liang P., Ge W.W. Association of telomere length with authentic pluripotency of ES/iPS cells. Cell Res. 2011;21:779–792. doi: 10.1038/cr.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Liang P., Liu D., Huang J., Songyang Z. Telomere regulation in pluripotent stem cells. Protein Cell. 2014;5:194–202. doi: 10.1007/s13238-014-0028-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D., Maehr R., Guo W., Eijkelenboom A., Snitow M., Chen A.E., Melton D.A. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D., Osafune K., Maehr R., Guo W., Eijkelenboom A., Chen S., Muhlestein W., Melton D.A. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- Ichida J.K., Blanchard J., Lam K., Son E.Y., Chung J.E., Egli D., Loh K.M., Carter A.C., Di Giorgio F.P., Koszka K. A small-molecule inhibitor of Tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Lv W., Ye X., Wang L., Zhang M., Yang H., Okuka M., Zhou C., Zhang X., Liu L. Zscan4 promotes genomic stability during reprogramming and dramatically improves the quality of iPS cells as demonstrated by tetraploid complementation. Cell Res. 2013;23:92–106. doi: 10.1038/cr.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang Q., Yin X., Yang W., Du Y., Hou P., Ge J., Liu C., Zhang W., Zhang X. Generation of iPSCs from mouse fibroblasts with a single gene, Oct4, and small molecules. Cell Res. 2011;21:196–204. doi: 10.1038/cr.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li K., Wei W., Ding S. Chemical approaches to stem cell biology and therapeutics. Cell Stem Cell. 2013;13:270–283. doi: 10.1016/j.stem.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. Linking telomere regulation to stem cell pluripotency. Trends Genet. 2017;33:16–33. doi: 10.1016/j.tig.2016.10.007. [DOI] [PubMed] [Google Scholar]
- Liu L., Bailey S.M., Okuka M., Munoz P., Li C., Zhou L., Wu C., Czerwiec E., Sandler L., Seyfang A. Telomere lengthening early in development. Nat. Cell Biol. 2007;9:1436–1441. doi: 10.1038/ncb1664. [DOI] [PubMed] [Google Scholar]
- Londono-Vallejo J.A., Der-Sarkissian H., Cazes L., Bacchetti S., Reddel R.R. Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res. 2004;64:2324–2327. doi: 10.1158/0008-5472.can-03-4035. [DOI] [PubMed] [Google Scholar]
- Long Y., Wang M., Gu H., Xie X. Bromodeoxyuridine promotes full-chemical induction of mouse pluripotent stem cells. Cell Res. 2015;25:1171–1174. doi: 10.1038/cr.2015.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan T.S., Gifford W.D., Driscoll S., Lettieri K., Rowe H.M., Bonanomi D., Firth A., Singer O., Trono D., Pfaff S.L. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N., Hochedlinger K. Guidelines and techniques for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;3:595–605. doi: 10.1016/j.stem.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Maherali N., Sridharan R., Xie W., Utikal J., Eminli S., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Marion R.M., Strati K., Li H., Tejera A., Schoeftner S., Ortega S., Serrano M., Blasco M.A. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4:141–154. doi: 10.1016/j.stem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Miyanari Y., Torres-Padilla M.E. Control of ground-state pluripotency by allelic regulation of Nanog. Nature. 2012;483:470–473. doi: 10.1038/nature10807. [DOI] [PubMed] [Google Scholar]
- Nichols J., Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Palm W., de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Pucci F., Gardano L., Harrington L. Short telomeres in ESCs lead to unstable differentiation. Cell Stem Cell. 2013;12:479–486. doi: 10.1016/j.stem.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R.P., Garrobo I., Foronda M., Palacios J.A., Marion R.M., Flores I., Ortega S., Blasco M.A. TRF1 is a stem cell marker and is essential for the generation of induced pluripotent stem cells. Nat. Commun. 2013;4:1946. doi: 10.1038/ncomms2946. [DOI] [PubMed] [Google Scholar]
- Shi Y., Desponts C., Do J.T., Hahm H.S., Scholer H.R., Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Silva J., Nichols J., Theunissen T.W., Guo G., van Oosten A.L., Barrandon O., Wray J., Yamanaka S., Chambers I., Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takai H., Smogorzewska A., de Lange T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- Tan M., Luo H., Lee S., Jin F., Yang J.S., Montellier E., Buchou T., Cheng Z., Rousseaux S., Rajagopal N. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Barbacioru C., Bao S., Lee C., Nordman E., Wang X., Lao K., Surani M.A. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-seq analysis. Cell Stem Cell. 2010;6:468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichroeb J.H., Kim J., Betts D.H. The role of telomeres and telomerase reverse transcriptase isoforms in pluripotency induction and maintenance. RNA Biol. 2016;13:707–719. doi: 10.1080/15476286.2015.1134413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valamehr B., Robinson M., Abujarour R., Rezner B., Vranceanu F., Le T., Medcalf A., Lee T.T., Fitch M., Robbins D. Platform for induction and maintenance of transgene-free hiPSCs resembling ground state pluripotent stem cells. Stem Cell Reports. 2014;2:366–381. doi: 10.1016/j.stemcr.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Yin Y., Ye X., Liu K., Zhu H., Wang L., Chiourea M., Okuka M., Ji G., Dan J. Molecular insights into the heterogeneity of telomere reprogramming in induced pluripotent stem cells. Cell Res. 2012;22:757–768. doi: 10.1038/cr.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Liu X., Chen J., Gao S., Lu L., Zhang H., Ding G., Wang Z., Chen Z., Shi T. Class I histone deacetylases are major histone decrotonylases: evidence for critical and broad function of histone crotonylation in transcription. Cell Res. 2017;27:898–915. doi: 10.1038/cr.2017.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Mao A., Tang B., Zeng Q., Gao S., Liu X., Lu L., Li W., Du J.X., Li J. Large-scale identification of protein crotonylation reveals its role in multiple cellular functions. J. Proteome Res. 2017;16:1743–1752. doi: 10.1021/acs.jproteome.7b00012. [DOI] [PubMed] [Google Scholar]
- Wernig M., Meissner A., Foreman R., Brambrink T., Ku M., Hochedlinger K., Bernstein B.E., Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Yeom Y.I., Fuhrmann G., Ovitt C.E., Brehm A., Ohbo K., Gross M., Hubner K., Scholer H.R. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- Zalzman M., Falco G., Sharova L.V., Nishiyama A., Thomas M., Lee S.L., Stagg C.A., Hoang H.G., Yang H.T., Indig F.E. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010;464:858–863. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Dan J., Wang H., Guo R., Mao J., Fu H., Wei X., Liu L. Tcstv1 and Tcstv3 elongate telomeres of mouse ES cells. Sci. Rep. 2016;6:19852. doi: 10.1038/srep19852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.Y., Li W., Lv Z., Liu L., Tong M., Hai T., Hao J., Guo C.L., Ma Q.W., Wang L. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Zhao T., Guan J., Zhang X., Fu Y., Ye J., Zhu J., Meng G., Ge J., Yang S. A XEN-like state bridges somatic cells to pluripotency during chemical reprogramming. Cell. 2015;163:1678–1691. doi: 10.1016/j.cell.2015.11.017. [DOI] [PubMed] [Google Scholar]
- Zhu S., Li W., Zhou H., Wei W., Ambasudhan R., Lin T., Kim J., Zhang K., Ding S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7:651–655. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.