Summary
Adult and embryonic stem cells exhibit fluctuating gene expression; however, the biological significance of stem cell heterogeneity is not well understood. We show that, in Drosophila, female germline stem cells (GSCs) exhibit heterogeneous expression of a GSC differentiation-promoting factor Regena (Rga). The Drosophila homolog of human SON, dsn, is required to maintain GSC heterogeneity by suppressing sustained high levels of Rga. Reducing the expression of Rga in dsn mutants restores GSC heterogeneity and self-renewal. Thus, GSC heterogeneity is linked to GSC homeostasis.
Keywords: Drosophila, germline stem cell, Regena, homeostasis, heterogeneity, CG8273, Dsn, self-renewal, differentiation
Graphical Abstract

Highlights
-
•
Female germline stem cells have heterogeneous Rga expression
-
•
dsn suppresses rga transcription and maintains heterogeneity
-
•
dsn maintains germline stem cells
-
•
Lowering rga restores heterogeneity and suppresses dsn phenotypes
Pek and colleagues report that the expression of differentiation factor Rga is heterogeneous between germline stem cells (GSCs) in the Drosophila ovaries. Dsn (human homolog of SON) represses rga transcription and promotes GSC heterogeneity, which is important for GSC homeostasis.
Introduction
In vertebrates and invertebrates, stem cells have been shown to exhibit heterogeneous gene expression, which may be important to regulate self-renewal and differentiation into distinct lineages (Chang et al., 2008, Graf and Stadtfeld, 2008, Imayoshi et al., 2013, Kobayashi et al., 2009, MacArthur et al., 2012, Ohlstein and Spradling, 2007). Drosophila female germline stem cell (GSC) niche represents a simple and tractable in vivo model with two to three GSCs that can be identified unambiguously by their location next to the cap cells and the presence of spherical spectrosomes (Figure 1A). Although extensively used as a stem cell model, heterogeneity in gene expression has never been reported in Drosophila female GSCs (Lehmann, 2012, Losick et al., 2011). Here, we report that the expression of a differentiation-promoting factor Regena (Rga) is heterogeneous between GSCs in the Drosophila ovaries. Dsn (human homolog of SON) represses rga transcription and promotes GSC heterogeneity, which is important for GSC homeostasis.
Figure 1.
Drosophila Female GSCs Exhibit Heterogeneous Rga Expression
(A) Drawing of a germarium showing the different cell types. TF, terminal filament; CC, cap cells; GSC, germline stem cell; CB, cytoblast; EC, escort cell.
(B) The rga-sisR-1 feedback loop. Transcription of rga pre-mRNA produces both sisR-1 (from the intron) and rga mRNA that is translated into Rga protein. sisR-1 represses rga transcription.
(C) Confocal images of a wild-type germarium stained for Rga (green) and Vasa (red). GSC #1 expresses higher level of Rga than GSC #2. Inset: magnification of GSCs #1 and #2. Scale bar: 10 μm.
(D) Chart showing the relative intensities of Rga and Vasa in GSCs #1 and #2 in (C).
(E) Chart showing the ratio of Rga intensities between GSCs in individual germaria.
(F) Chart showing the percentage of germaria with and without heterogeneous expression of Rga as depicted in (E) where cutoff was set at 1.5. n = 20 germaria.
(G) Diagram showing the scoring of cell cycle stages (G1/S and G2) according to fusome morphology.
(H) Chart showing percentages of GSCs with high, low, or homogeneous levels of Rga at G1/S or G2 stages. N = 37–50. ∗p < 0.05, two-tailed Z test. ns, p > 0.05.
(I) Chart showing percentages of germaria with GSC pairs having heterogeneous or homogeneous levels of Rga at different combinations of cell cycle stages (G1/S-G1/S, G1/S-G2, or G2-G2). N = 25–37. ∗p < 0.05, two-tailed Z test. ns, p > 0.05.
(J and K) Confocal images depicting (J) two GSCs having heterogeneous Rga expression at G2, and (K) two GSCs having homogeneous Rga expression at G1/S.
Results
Drosophila Female GSCs Exhibit Heterogeneous Rga Expression
Rga (NOT2 in the CCR4-NOT deadenylase complex) was reported to promote GSC differentiation, where high and low levels of Rga promote and inhibit GSC differentiation respectively (Wong et al., 2017). The expression of rga is being regulated by a negative feedback loop via the sisR-1 noncoding RNA (Figure 1B) (Osman et al., 2016, Pek, 2018, Pek et al., 2015). sisR-1 belongs to a class of stable intronic sequence RNAs that frequently regulate gene expression via feedback loops (Pek, 2018, Pek and Okamura, 2015). As previous efforts to visualize rga transcripts by in situ hybridization were unsuccessful (Wong et al., 2017), we began our study by examining the expression pattern of Rga protein in GSCs using a specific antibody against Rga (Temme et al., 2004, Wong et al., 2017). Unlike Vasa (a germline marker) that was expressed at similar levels between adjacent GSCs, Rga expression was highly heterogeneous (Figures 1C and 1D). Within the same niche, one GSC (GSC #1) expresses a higher level of Rga compared with an adjacent GSC (GSC #2), while the expression of Vasa was similar (Figures 1C and 1D). We characterized the extent of heterogeneity by measuring the levels of Rga in adjacent GSCs and calculated the ratio (see Experimental Procedures). GSCs ranged from homogeneous (ratio of 1) to highly heterogeneous (ratio of ∼4.5) (Figure 1E). A ratio of 1.5 was sufficient to be scored as heterogeneous under visual inspection. Using this criterion, we observed that the expression of Rga was heterogeneous between adjacent GSCs in ∼65% of the germaria examined (Figure 1F).
Heterogeneity appears to be specific to Rga as the expression of another component of the deadenylase complex CCR4 did not exhibit heterogeneity between GSCs (Figure S1A). To further investigate if heterogeneity correlates with the cell cycle, we examined the different stages of cell division using fusome morphology as a marker (Figure 1G) (de Cuevas and Spradling, 1998). Consistent with our earlier analysis, ∼40% of the germaria exhibited homogeneous Rga expression (Figure S1B). Approximately 30% of the GSCs were at G1/S phase, while the remaining ∼70% were at G2 phase, which is similar to what were reported in previous studies (Figure S1C) (Hsu et al., 2008, Morris and Spradling, 2011). Interestingly, when we examined the GSCs that were heterogeneous (either high or low Rga), most (>70%) of them were at G2 phase (Figure 1H); however, this was not the case for GSCs expressing homogeneous levels of Rga (Figure 1H). Furthermore, neighboring GSCs both at G2 were more heterogeneous, while those at G1/S were homogeneous (Figures 1I–1K and S1D). Therefore, our data suggest that the heterogeneity of Rga correlates with the cell cycle, with GSCs in G2 being the most heterogeneous.
dsn Represses rga Expression and Maintains Rga Heterogeneity in GSCs
In a screen for nuclear double-stranded RNA binding proteins that regulate sisR-1, we previously identified CG8273 as a candidate gene (Wong et al., 2017). CG8273 (called dSon or dsn hereafter) is highly expressed in the ovaries (Figure S2). dsn is the homolog of human SON, which is involved in transcriptional repression and splicing (Kim et al., 2016a, Lu et al., 2014, Sun et al., 2001). SON had been shown to regulate self-renewal in human embryonic stem cells and mutations in SON are associated with abnormal brain development and leukemia (Kim et al., 2016a, Kim et al., 2016b, Lu et al., 2013, Tokita et al., 2016).
To examine the function of dsn, we used a strain containing a transposon insertion at the 5′UTR of dsn, which dramatically reduced the expression of dsn mRNA (Figures 2A and 2B). In dsn homozygous mutant ovaries, the steady-state level of sisR-1 was upregulated (Figure 2C). Since SON had been shown to be a transcriptional repressor and splicing enhancer, we asked if dsn regulates rga pre-mRNA (sisR-1 precursor) expression and/or splicing. Consistent with a role in repressing rga transcription, rga pre-mRNA levels were also upregulated ∼2.5-fold in dsn mutant ovaries compared with controls (Figure 2D). In dsn mutant ovaries, splicing of rga pre-mRNA was normal, as shown by RT-PCR using primers flanking the intron, and the levels of rga mRNA also increased in a similar magnitude of ∼3-fold as rga pre-mRNA (Figures 2E and 2F). Taken together, we conclude that dsn primarily represses the transcription of rga pre-mRNA.
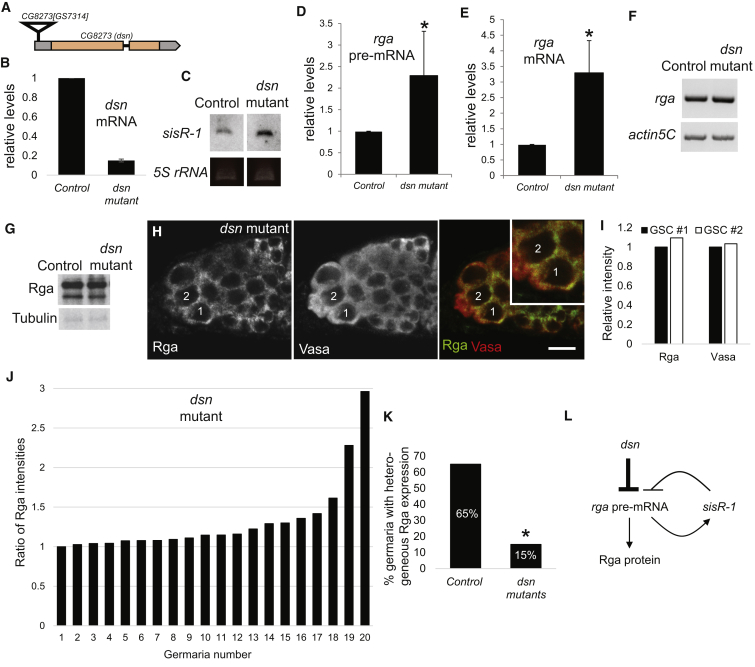
Figure 2.
dsn Represses rga Expression and Maintains Rga Heterogeneity in GSCs
(A) Gene locus showing the position of the transposon insertion GS7314 in the dsn locus at the 5′UTR.
(B) qPCR showing the relative levels of dsn in y w control and dsn homozygous mutant ovaries. Actin5C mRNA was used as a loading control.
(C) A representative northern blot showing the abundance of sisR-1 in control and dsn mutant ovaries. 5S rRNA was used as a loading control.
(D and E) Charts showing the relative abundance of rga (D) pre-mRNA and (E) mRNA in control and dsn mutant ovaries. N = 3 biological replicates. Error bars represent SD. ∗p < 0.01, two-tailed t test.
(F) Gel showing RT-PCR of rga mRNA in control and dsn mutant ovaries showing correct splicing. Actin5C was used as a loading control.
(G) Western blot showing the abundance of Rga protein in control and dsn mutant ovaries. Tubulin was used as a loading control.
(H) Confocal images of a dsn mutant germarium stained for Rga (green) and Vasa (red). GSC #1 expresses similar level of Rga with GSC #2. Inset: magnification of GSCs #1 and #2. Scale bar: 10 μm.
(I) Chart showing the relative intensities of Rga and Vasa in GSCs #1 and #2 in (H).
(J) Chart showing the ratio of Rga intensities between GSCs in individual germaria in dsn mutants.
(K) Chart showing the percentages of germaria with heterogeneous expression of Rga in GSCs in control and dsn mutant ovaries. N = 20. ∗p = 0.001, two-tailed Z test.
(L) Model showing Dsn directly represses the expression of rga pre-mRNA independent of the sisR-1-mediated negative feedback loop.
While we observed an increase in rga mRNA levels in dsn mutant ovaries, we did not detect an obvious change in Rga protein expression by western blotting (Figure 2G), suggesting additional feedback mechanism(s) that regulate Rga translation and/or protein stability during late oogenesis. Nevertheless, at the cellular level, we observed that, in ∼85% of dsn mutant ovarioles, GSCs exhibited sustained high levels of Rga expression leading to a loss of heterogeneity in Rga staining (Figures 2H–2K). Adjacent GSCs (#1 and #2) now expressed similar levels of Rga, a pattern similar to that of Vasa (Figures 2H–2J). This led to a drop in the percentage of germaria with GSCs having heterogeneous Rga expression from ∼65% in controls to ∼15% in dsn mutants (Figure 2K, p = 0.00124, two-tail Z test). We conclude that dsn is required to maintain GSC heterogeneity by repressing the expression of rga pre-mRNA (Figure 2L). We suggest that, in dsn mutants, rga is upregulated leading to a higher level and perdurance of Rga protein in the GSCs, and hence loss of heterogeneity.
dsn Maintains GSC Heterogeneity and Self-Renewal by Repressing rga
The tuning of Rga had been previously found to regulate GSC self-renewal (Wong et al., 2017, Yan et al., 2014). Overexpression of Rga in GSCs promotes differentiation, while knockdown of Rga promotes self-renewal (Wong et al., 2017). Since Rga was upregulated in dsn mutant GSCs, we examined if dsn mutants displayed any GSC maintenance defects. We found that day 2 dsn mutant females laid significantly fewer eggs compared with controls (Figure 3A; control, ∼53 ± 11 eggs/female/day versus dsn mutant, ∼36 ± 5 eggs/female/day; p = 0.0003, two-tailed t test). However, the eggs hatched with similar rates as controls (Figure S3A), consistent with an unperturbed expression of Rga protein in the whole ovaries assayed by western blotting (Figure 2G). At day 21, the egg laying phenotype became more severe (Figure 3A; control, ∼60 ± 14 eggs/female/day versus dsn mutant, ∼20 ± 4 eggs/female/day; p < 0.0001, two-tailed t test), suggesting a GSC defect. Closer examination of the GSCs revealed a significant decrease in GSC number in dsn mutant ovaries relative to controls (Figures 3B and 3C; control, ∼2.4 ± 0.7 GSCs versus dsn mutant, ∼1.2 ± 0.8 GSCs; p = 0.00006, two-tailed t test). The GSC phenotype worsened with age (Figures 3B and 3C), confirming a GSC maintenance defect. Analysis of dsn/Df(3R)Exel6153 showed a similar GSC loss phenotype (Figures 3D and 3E; control, ∼3.0 ± 1.0 GSCs versus dsn/Df, ∼1.6 ± 1.5 GSCs; p = 0.04, two-tailed t test). Knockdown of dsn in the germline by a germline-specific driver vasa-Gal4 also led to GSC loss (Figures 3F, 3G, and S3B; control, ∼2.5 ± 0.9 GSCs versus dsn RNAi, ∼1.7 ± 0.9 GSCs; p = 0.02, two-tailed t test), confirming a role of dsn in promoting GSC self-renewal cell autonomously.
Figure 3.
dsn Maintains GSCs
(A) Chart showing the number of eggs laid per female per day in control and dsn mutants at day 2 and 21 post eclosure. N = 6–10 experiments. ∗p < 0.001, two-tailed t test.
(B) Chart showing the number of GSCs in control and dsn mutant ovaries at day 2 and 21 post eclosure. N = 10–20. ∗p < 0.001, two-tailed t test.
(C) Confocal images showing control and dsn mutant germaria at day 2 and 21 post eclosure stained with alpha-spectrin (green) and Vasa (red).
(D) Chart showing the number of GSCs in control and dsn/Df ovaries. N = 10. ∗p < 0.05, two-tailed t test.
(E) Confocal images showing control and dsn/Df germaria stained with alpha-spectrin (green) and Vasa (red).
(F) Chart showing the number of GSCs in control and dsn RNAi ovaries. N = 20. ∗p < 0.02, two-tailed t test.
(G) Confocal images showing control and dsn RNAi germaria stained with alpha-spectrin (green) and Vasa (red).
∗GSCs. Error bars represent standard deviation. Scale bar: 10 μm.
To investigate if the GSC loss phenotype in dsn mutants was due to sustained high levels of Rga in the GSCs, we reduced the dosage of Rga by removing a copy of rga in the dsn RNAi background. In dsn RNAi ovaries, 12.5% of germaria had GSCs with Rga heterogeneity (Figures 4A–4D). When we reduced the dosage of Rga by 50% in the dsn RNAi ovaries, Rga heterogeneity in GSCs reverted to normal in 56% of the germaria examined (Figures 4A–4D; dsn RNAi, 12.5% versus dsn RNAi; rga/+, 56%; p = 0.01, two-tail Z test). Moreover, these flies had a normal number of GSCs compared with dsn RNAi (Figures 4E and 4F; dsn RNAi, ∼1.5 ± 0.9 GSCs versus dsn RNAi; rga/+, ∼2.3 ± 0.9 GSCs; p = 0.01, two-tailed t test). Consistently, egg laying was also rescued back to normal (Figure 4G; control, ∼70 ± 35 eggs/female/day versus dsn RNAi, ∼28 ± 24 eggs/female/day versus dsn RNAi; rga/+, ∼117 ± 47 eggs/female/day; p < 0.0001, two-tailed t test). Taken together, our genetic interaction analysis confirmed that dsn regulates GSC heterogeneity and self-renewal by suppressing high levels of rga in the GSCs.
Figure 4.
Loss of GSCs in dsn RNAi Is Due to High Levels of Rga
(A) Confocal images of dsn RNAi and dsn RNAi; rga/+ germaria stained for Rga (green) and Vasa (red). Adjacent GSCs were labeled GSC #1 and GSC #2. Inset: magnification of GSCs #1 and #2. Scale bar: 10 μm.
(B) Chart showing the relative intensities of Rga and Vasa in GSCs #1 and #2 in (A).
(C) Chart showing the ratio of Rga intensities between GSCs in individual germaria in dsn RNAi and dsn RNAi; rga/+ ovaries.
(D) Chart showing the percentages of germaria with heterogeneous expression of Rga in GSCs in dsn RNAi and dsn RNAi; rga/+ ovaries. N = 16. ∗p = 0.009, two-tailed Z test.
(E) Chart showing the number of GSCs in dsn RNAi and dsn RNAi; rga/+ ovaries. N = 20. ∗p < 0.02, two-tailed t test.
(F) Confocal images showing dsn RNAi and dsn RNAi; rga/+ germaria stained with alpha-spectrin (green) and Vasa (red). ∗GSCs. Scale bar: 10 μm.
(G) Chart showing the number of eggs laid per female per day in control, dsn RNAi, and dsn RNAi; rga/+ females. N = 6–12 experiments. ∗p < 0.001, two-tailed t test. ns, p > 0.05.
(H) Diagrammatic representation of how Rga heterogeneity can regulate GSC self-renewal versus differentiation.
Error bars represent standard deviation.
Discussion
We suggest that GSCs exhibit heterogeneous levels of Rga to coordinate self-renewal and differentiation. Prolonged overexpression of Rga triggers GSC differentiation while low Rga expression promotes GSC self-renewal (Wong et al., 2017). We envision that, at a given point of time, adjacent GSCs express different levels of Rga and are thus transiently primed toward differentiation (Figure 4H). This allows a GSC to respond rapidly and appropriately to changes in niche signals in response to changes in the environment. Since both GSCs do not respond equally, such a mechanism also creates a buffer against random fluctuations of differentiation-promoting signals that can result in unwanted loss of GSCs. As G2 in GSCs is highly regulated by the insulin and TOR pathways (Hsu et al., 2008, LaFever et al., 2010), it may serve as a critical window when signaling pathways intersect with Rga to produce a graded response of self-renewal versus differentiation.
Stem cell heterogeneity has been reported and characterized in mammalian embryonic and adult stem cells such as the spermatogonial, hematopoietic, intestinal, and epithelial stem cells (Donati and Watt, 2015, Goodell et al., 2015, Graf and Stadtfeld, 2008, Krieger and Simons, 2015). Mathematical modeling had also provided support for a role of stem cell heterogeneity in balancing self-renewal and differentiation to achieve tissue homeostasis (Greulich and Simons, 2016). Our study suggests that the phenomenon of stem cell heterogeneity is also conserved in invertebrate adult stem cells. Importantly, it provides a simple model system to investigate the role of stem cell heterogeneity in tissue homeostasis.
How such Rga heterogeneity is achieved is not understood. Negative feedback loops have been proposed in other systems to generate oscillatory gene expression (Imayoshi et al., 2013, Isomura and Kageyama, 2014, Kobayashi et al., 2009, MacArthur et al., 2012). One possibility is that an oscillatory mode of Rga expression is generated by the rga-sisR-1 negative feedback loop (Pek, 2018). The other possibilities are that Rga can fluctuate in a random manner or in response to some unknown factors during G2. In future, it would be important to understand the dynamics of Rga in GSCs. This can be achieved by long-term live imaging and quantitative analyses of the kinetics of translation and protein degradation at the single-cell level.
Experimental Procedures
A detailed description of all methods is included in the Supplemental Information.
Fly Strains
y w flies were used a controls unless otherwise stated. The following strains were used in this study: CG8273GS7314 (Kyoto #201169), Df(3R)6153 (Bloomington #7632), CG8273 RNAi (TRiP HMS00114 Bloomington #34805), MTD-Gal4 (Petrella et al., 2007), and vasa-Gal4 (kind gift from Y. Yamashita). Flies were maintained at 25°C, while the RNAi flies at 29°C. Newly eclosed flies were fed with wet yeast paste for 2 to 21 days before dissection.
Immunostaining
Immunostaining was perform as previously described (Pek and Kai, 2011, Pek et al., 2012, Wong et al., 2017). Ovaries were fixed in a solution containing 16% paraformaldehyde and Grace's medium (2:1 ratio) for 20 min. They were then rinsed and washed in PBX (PBS containing 0.2% Triton X-100), and pre-absorbed in PBX with 5% normal goat serum. Incubation in primary antibodies was done at room temperature overnight. The next day, ovaries were washed in PBX and incubated in secondary antibodies for 4 hr at room temperature. Finally, the ovaries were washed in PBX before mounting on slides. The primary antibodies used were rabbit anti-Rga (1:500, kind gift from E. Wahle), guinea pig anti-Vasa (1:1,000, kind gift from T. Kai), mouse anti-alpha-spectrin (1:2, Developmental Studies Hybridoma Bank), and rabbit anti-CCR4 (1:500, kind gift from E. Wahle). Specificity of Rga antibody was verified previously (Wong et al., 2017). Images were taken using the Carl Zeiss LSM 5 Exciter Upright confocal microscope. Signal intensities were quantified using ImageJ software. Images were processed using Adobe Photoshop software.
Identification and Scoring of GSCs
GSCs were identified based on their location and size (Morris and Spradling, 2011) or the presence of spectrosomes. Germaria containing two GSCs clearly visible on the same confocal plane were randomly selected and Rga intensity was measured using ImageJ software. The ratios of Rga intensity of neighboring GSCs were calculated using the GSC with the lower Rga level as the denominator. Therefore, GSCs that are homogeneous will have a ratio close to 1, while those that are heterogeneous will be greater than 1. We used a cutoff of 1.5 to determine if GSCs are heterogeneous or not, because a ratio of 1.5 was sufficient to determine heterogeneity by visual inspection.
Author Contributions
A.Y.E.N. performed northern blotting, RT-PCR, and qPCR. K.R.G.P. performed qPCR, western blotting, and fertility tests. J.W.P. performed the remaining experiments.
Acknowledgments
We thank E. Wahle, T. Kai, Y. Yamashita, Developmental Studies Hybridoma Bank, Kyoto Stock Center, and the Bloomington Stock Center for reagents; and members of the Pek laboratory I. Osman and M. Tay for discussion. The authors are supported by the Temasek Life Sciences Laboratory.
Published: June 7, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and three figures and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.05.005.
Supplemental Information
References
- Chang H.H., Hemberg M., Barahona M., Ingber D.E., Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cuevas M., Spradling A.C. Morphogenesis of the Drosophila fusome and its implications for oocyte specification. Development. 1998;125:2781–2789. doi: 10.1242/dev.125.15.2781. [DOI] [PubMed] [Google Scholar]
- Donati G., Watt F.M. Stem cell heterogeneity and plasticity in epithelia. Cell Stem Cell. 2015;16:465–476. doi: 10.1016/j.stem.2015.04.014. [DOI] [PubMed] [Google Scholar]
- Goodell M.A., Nguyen H., Shroyer N. Somatic stem cell heterogeneity: diversity in the blood, skin and intestinal stem cell compartments. Nat. Rev. Mol. Cell Biol. 2015;16:299–309. doi: 10.1038/nrm3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T., Stadtfeld M. Heterogeneity of embryonic and adult stem cells. Cell Stem Cell. 2008;3:480–483. doi: 10.1016/j.stem.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Greulich P., Simons B.D. Dynamic heterogeneity as a strategy of stem cell self-renewal. Proc. Natl. Acad. Sci. USA. 2016;113:7509–7514. doi: 10.1073/pnas.1602779113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H.J., LaFever L., Drummond-Barbosa D. Diet controls normal and tumorous germline stem cells via insulin-dependent and -independent mechanisms in Drosophila. Dev. Biol. 2008;313:700–712. doi: 10.1016/j.ydbio.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I., Isomura A., Harima Y., Kawaguchi K., Kori H., Miyachi H., Fujiwara T., Ishidate F., Kageyama R. Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science. 2013;342:1203–1208. doi: 10.1126/science.1242366. [DOI] [PubMed] [Google Scholar]
- Isomura A., Kageyama R. Ultradian oscillations and pulses: coordinating cellular responses and cell fate decisions. Development. 2014;141:3627–3636. doi: 10.1242/dev.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Baddoo M.C., Park E.Y., Stone J.K., Park H., Butler T.W., Huang G., Yan X., Pauli-Behn F., Myers R.M. SON and its alternatively spliced isoforms control MLL complex-mediated H3K4me3 and transcription of leukemia-associated genes. Mol. Cell. 2016;61:859–873. doi: 10.1016/j.molcel.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Shinde D.N., Reijnders M.R.F., Hauser N.S., Belmonte R.L., Wilson G.R., Bosch D.G.M., Bubulya P.A., Shashi V., Petrovski S. De novo mutations in SON disrupt RNA splicing of genes essential for brain development and metabolism, causing an intellectual-disability syndrome. Am. J. Hum. Genet. 2016;99:711–719. doi: 10.1016/j.ajhg.2016.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Mizuno H., Imayoshi I., Furusawa C., Shirahige K., Kageyama R. The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev. 2009;23:1870–1875. doi: 10.1101/gad.1823109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger T., Simons B.D. Dynamic stem cell heterogeneity. Development. 2015;142:1396–1406. doi: 10.1242/dev.101063. [DOI] [PubMed] [Google Scholar]
- LaFever L., Feoktistov A., Hsu H.J., Drummond-Barbosa D. Specific roles of target of rapamycin in the control of stem cells and their progeny in the Drosophila ovary. Development. 2010;137:2117–2126. doi: 10.1242/dev.050351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R. Germline stem cells: origin and destiny. Cell Stem Cell. 2012;10:729–739. doi: 10.1016/j.stem.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick V.P., Morris L.X., Fox D.T., Spradling A. Drosophila stem cell niches: a decade of discovery suggests a unified view of stem cell regulation. Dev. Cell. 2011;21:159–171. doi: 10.1016/j.devcel.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Goke J., Sachs F., Jacques P.E., Liang H., Feng B., Bourque G., Bubulya P.A., Ng H.H. SON connects the splicing-regulatory network with pluripotency in human embryonic stem cells. Nat. Cell Biol. 2013;15:1141–1152. doi: 10.1038/ncb2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Ng H.H., Bubulya P.A. The role of SON in splicing, development, and disease. Wiley Interdiscip. Rev. RNA. 2014;5:637–646. doi: 10.1002/wrna.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur B.D., Sevilla A., Lenz M., Muller F.J., Schuldt B.M., Schuppert A.A., Ridden S.J., Stumpf P.S., Fidalgo M., Ma'ayan A. Nanog-dependent feedback loops regulate murine embryonic stem cell heterogeneity. Nat. Cell Biol. 2012;14:1139–1147. doi: 10.1038/ncb2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris L.X., Spradling A.C. Long-term live imaging provides new insight into stem cell regulation and germline-soma coordination in the Drosophila ovary. Development. 2011;138:2207–2215. doi: 10.1242/dev.065508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B., Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science. 2007;315:988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- Osman I., Tay M.L., Pek J.W. Stable intronic sequence RNAs (sisRNAs): a new layer of gene regulation. Cell. Mol. Life Sci. 2016;73:3507–3519. doi: 10.1007/s00018-016-2256-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pek J.W. Stable intronic sequence RNAs engage in feedback loops. Trends Genet. 2018;34:330–332. doi: 10.1016/j.tig.2018.01.006. [DOI] [PubMed] [Google Scholar]
- Pek J.W., Kai T. A role for vasa in regulating mitotic chromosome condensation in Drosophila. Curr. Biol. 2011;21:39–44. doi: 10.1016/j.cub.2010.11.051. [DOI] [PubMed] [Google Scholar]
- Pek J.W., Ng B.F., Kai T. Polo-mediated phosphorylation of Maelstrom regulates oocyte determination during oogenesis in Drosophila. Development. 2012;139:4505–4513. doi: 10.1242/dev.082867. [DOI] [PubMed] [Google Scholar]
- Pek J.W., Okamura K. Regulatory RNAs discovered in unexpected places. Wiley Interdiscip. Rev. RNA. 2015;6:671–686. doi: 10.1002/wrna.1309. [DOI] [PubMed] [Google Scholar]
- Pek J.W., Osman I., Tay M.L., Zheng R.T. Stable intronic sequence RNAs have possible regulatory roles in Drosophila melanogaster. J. Cell Biol. 2015;211:243–251. doi: 10.1083/jcb.201507065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella L.N., Smith-Leiker T., Cooley L. The Ovhts polyprotein is cleaved to produce fusome and ring canal proteins required for Drosophila oogenesis. Development. 2007;134:703–712. doi: 10.1242/dev.02766. [DOI] [PubMed] [Google Scholar]
- Sun C.T., Lo W.Y., Wang I.H., Lo Y.H., Shiou S.R., Lai C.K., Ting L.P. Transcription repression of human hepatitis B virus genes by negative regulatory element-binding protein/SON. J. Biol. Chem. 2001;276:24059–24067. doi: 10.1074/jbc.M101330200. [DOI] [PubMed] [Google Scholar]
- Temme C., Zaessinger S., Meyer S., Simonelig M., Wahle E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 2004;23:2862–2871. doi: 10.1038/sj.emboj.7600273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokita M.J., Braxton A.A., Shao Y., Lewis A.M., Vincent M., Kury S., Besnard T., Isidor B., Latypova X., Bezieau S. De novo truncating variants in SON cause intellectual disability, congenital malformations, and failure to thrive. Am. J. Hum. Genet. 2016;99:720–727. doi: 10.1016/j.ajhg.2016.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J.T., Akhbar F., Ng A.Y.E., Tay M.L., Loi G.J.E., Pek J.W. DIP1 modulates stem cell homeostasis in Drosophila through regulation of sisR-1. Nat. Commun. 2017;8:759. doi: 10.1038/s41467-017-00684-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., Neumuller R.A., Buckner M., Ayers K., Li H., Hu Y., Yang-Zhou D., Pan L., Wang X., Kelley C. A regulatory network of Drosophila germline stem cell self-renewal. Dev. Cell. 2014;28:459–473. doi: 10.1016/j.devcel.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.