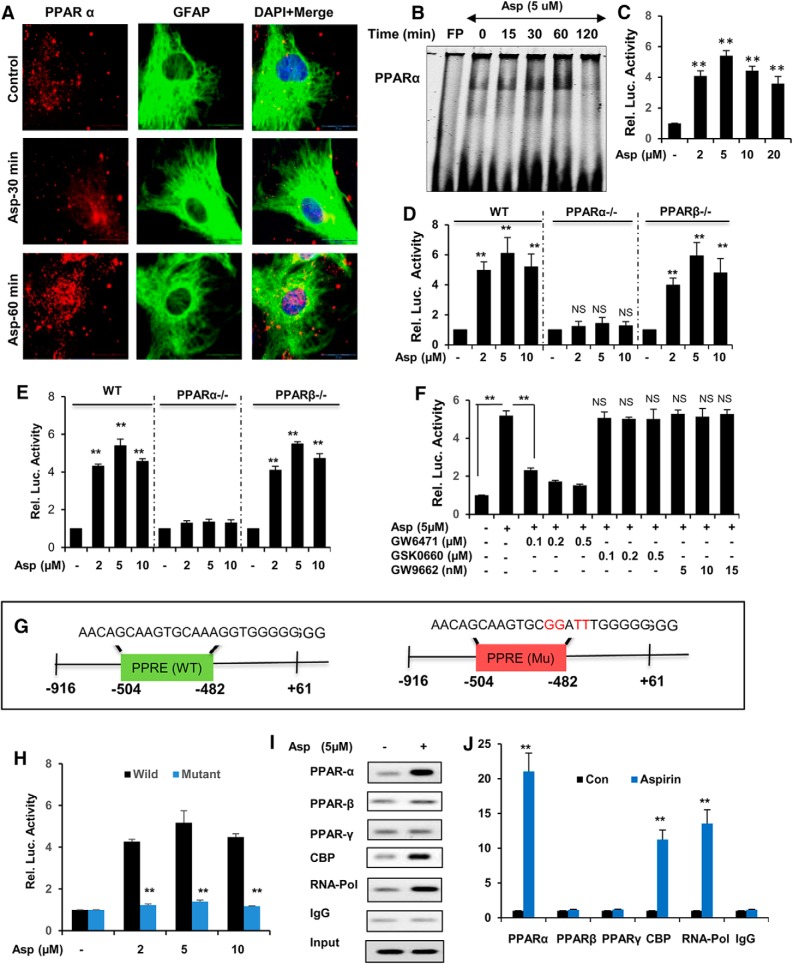
Figure 3.
Activation of PPARα by aspirin transcriptionally modulates TFEB expression. A, Primary astrocytes were treated with 5 μm aspirin for 30 min and 60 min, followed by double labeling for PPARα and GFAP. Scale bar, 20 μm. DAPI was used for staining nuclei. B, DNA-binding activity of PPARα after 15, 30, 60, and 120 min treatment of astrocytes with 5 μm aspirin as demonstrated by EMSA. FP, Free probe. C, WT astrocytes were transfected with pPPRE-luciferase construct for 24 h, followed by treatment with different doses of aspirin (2, 5, and 10 μm) for 8 h and were examined by luciferase assay. WT, Ppara-null, and Pparb-null astrocytes transfected with pPPRE-luciferase (D) or pTFEB(WT)-luciferase (E) construct for 24 h were treated with 2, 5, and 10 μm aspirin for 8 h and analyzed by luciferase assay. F, WT mouse astrocytes transfected with pTFEB(WT)-luciferase for 24 h were pretreated with different doses of PPARα antagonist GW6471 (100, 200, and 300 nm), PPARβ antagonist GSK0660 (100, 200, and 300 nm), and PPARγ antagonist GW9662 (5, 10, and 15 nm) for 1 h, followed by 5 μm aspirin treatment and subsequently subjected to luciferase assay. G, WT astrocytes transfected with pTFEB(WT)-luciferase or pTFEB(mutant)-luciferase construct for 24 h were treated with 5 μm aspirin for 8 h and analyzed by luciferase assay. H, Primary astrocytes treated with 5 μm aspirin for 2 h were analyzed for the recruitment of PPARα, PPARβ, PPARγ, CBP, and RNA polymerase to the conserved PPRE site on Tfeb promoter by performing ChIP, followed by semiquantitative RT-PCR (I) and quantitative RT-PCR (J). All values were normalized with input and are represented as fold change with respect to untreated controls. Data are shown as mean ± SD. IP, Immunoprecipitation; CBP, CREB binding protein. Statistical analysis was conducted using Student's t test; **p < 0.001.