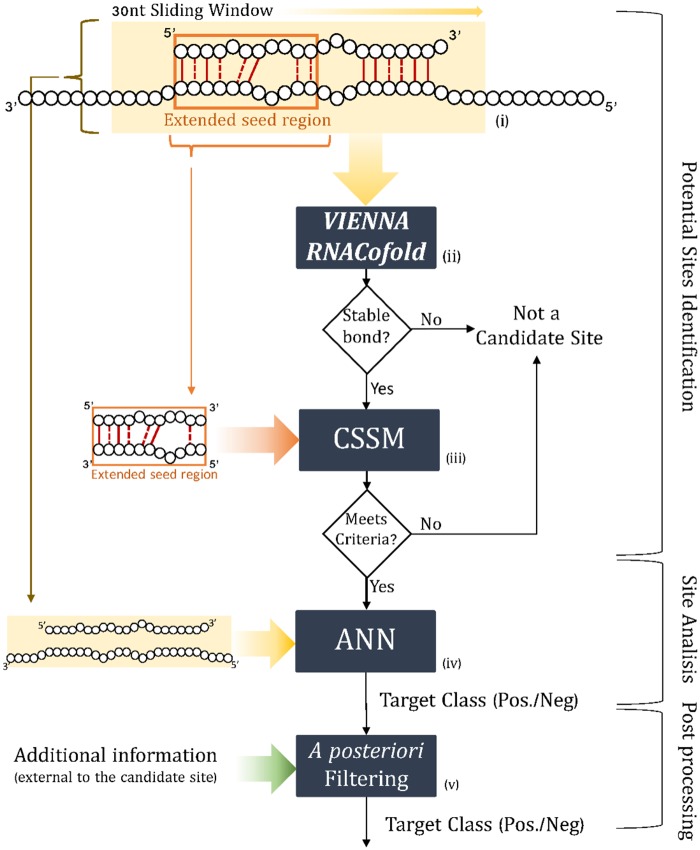
Fig 1. Schematic of the process used by miRAW to evaluate a miRNA binding site.
(i) A 30nt sliding window (with 5nt step) is used to scan the 3’UTR of a gene; (ii) The Vienna RNACofold software package is used to estimate whether the microRNA and the 30nt transcript can form a stable bond; (iii) If a stable bond is predicted, miRAW checks if the extended seed region meets the criteria defined in the candidate site selection method (CSSM); (iv) If the criteria are met, the full mature microRNA transcript and 30nt corresponding to the candidate site are fed into miRAW’s neural network to generate a classification; (v) The prediction can be refined by a filtering step that applies additional information that is external to the miRNA:site duplex.