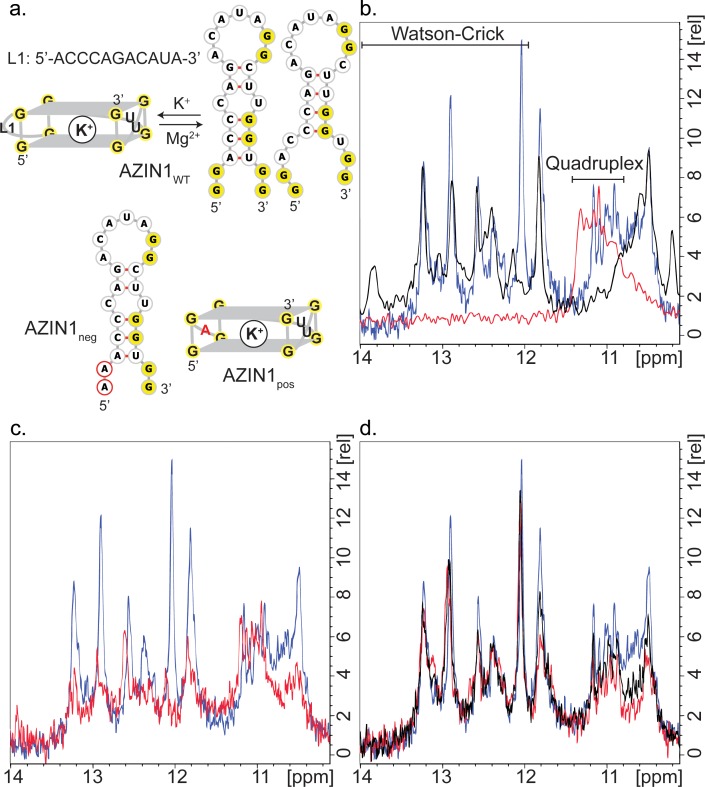
Figure 5. Structure of AZIN1wt.
(a) Derived model for the equilibrium between two possible hairpin conformers and the G2-quadruplex of AZIN1wt; K+ is expected to favor the G2-quadruplex; Na+ or Mg2+ are expected to favor the hairpin conformers, which are predicted by Mfold with free energies ΔG of −2.5 (left) and −1.8 kcal/mol (right). The mutants AZIN1neg and AZIN1pos were designed to stabilize the hairpin conformation and the G2-quadruplex, respectively: note the mutation of the first G2-tract at the 5’-end in AZIN1neg and the difference of the G2-quadruplex in the first loop from 5’-end (L1) for AZIN1pos. Sites of mutation are marked in red. (b) Overlay of 1H NMR spectra corresponding to the imino region of AZIN1wt (blue), AZIN1neg (black) and AZIN1pos (red) in 100 mM KCl (0.1 mM RNA). (c) Overlay of 1H NMR spectra corresponding to the imino region of AZINwt in 100 mM KCl (blue) and 200 mM KCl (red). (d) 1H NMR spectra after titration of spermine (Spm) to AZIN1wt in 100 mM KCl. Before addition of Spm (blue); after addition of Spm at RNA:Spm ratio of 1:1 (black) and 1:2 (red). Note the stronger decrease of the imino signals < 12 ppm.