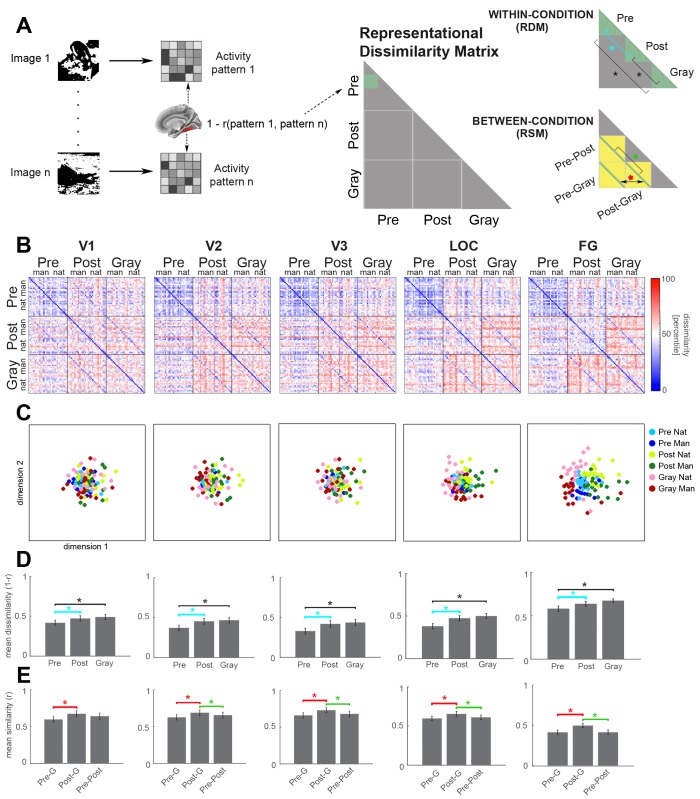
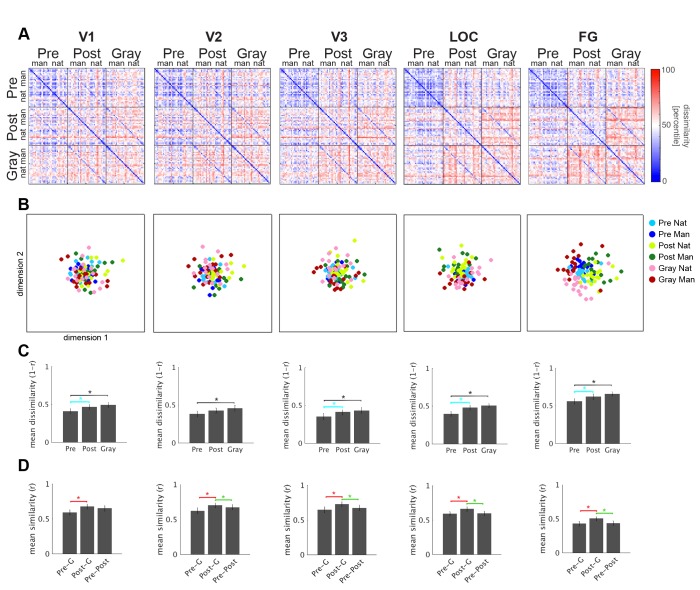
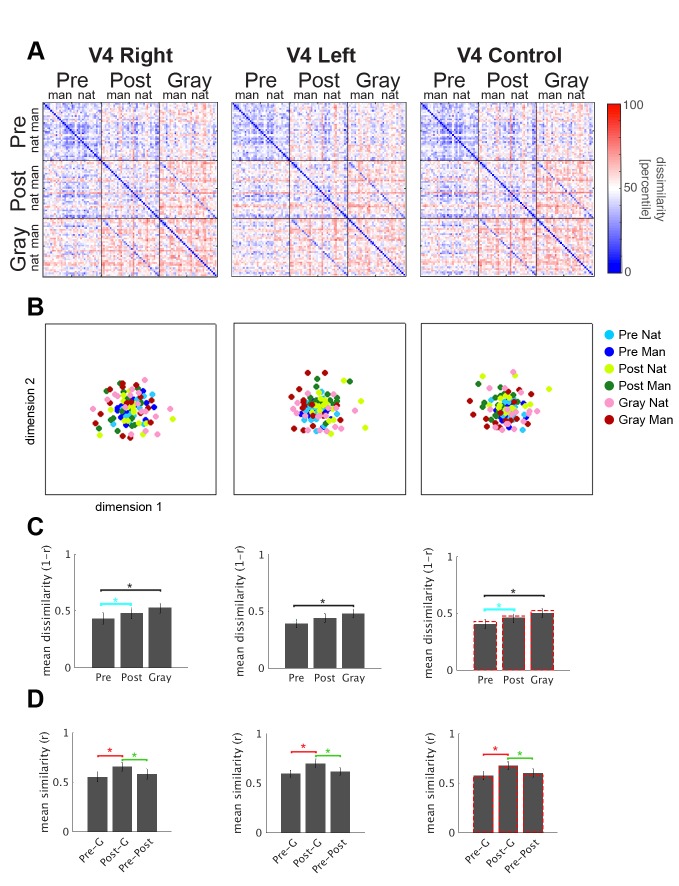
Figure 3. Neural representation format of individual images in visual regions.
(A) Analysis schematic. Dissimilarity (1 – Pearson’s r) between the neural response patterns to pairs of images was computed to construct the representational dissimilarity matrix (RDM) for each ROI. Two statistical analyses were performed: First (top-right panel, ‘within-condition’), mean dissimilarity across image pairs was calculated for each condition (green triangles), and compared between conditions (brackets). Cyan bracket highlights the main effect of interest (disambiguation). Second (bottom-right panel, ‘between-condition’), the mean of between-condition diagonals (green lines) was compared. For ease of interpretation, this analysis was carried out on the representational similarity matrix (RSM). Each element in the diagonal represents the neural similarity between the same Mooney image presented in different stages (Pre-Post), or between a Mooney image and its corresponding gray-scale image (Pre-Gray and Post-Gray). (B) Group-average RDMs for V1, V2, V3, LOC and FG ROIs in the right hemisphere. Black lines delimit boundaries of each condition. Within each condition, natural (‘nat’) and man-made (‘man’) images are grouped together. (C) 2-D MDS plots corresponding to the RDMs in B. Pre-disambiguation, post-disambiguation, and gray-scale images are shown as blue, yellow-green, and pink-red dots, respectively. (D) Mean within-condition representational dissimilarity between different images for each ROI, corresponding to the ‘within-condition’ analysis depicted in A. (E) Mean between-condition similarity for the same or corresponding images for each ROI, corresponding to the ‘between-condition’ analysis depicted in A. In D and E, asterisks denote significant differences (p<0.05, Wilcoxon signed-rank test, FDR-corrected), and error bars denote s.e.m. across subjects. Results from V4 are shown in Figure 3—figure supplement 2. Interactive 3-dimensional MDS plots corresponding to first-order RDMs for each ROI can be found at: https://gonzalezgarcia.github.io/mds.html.