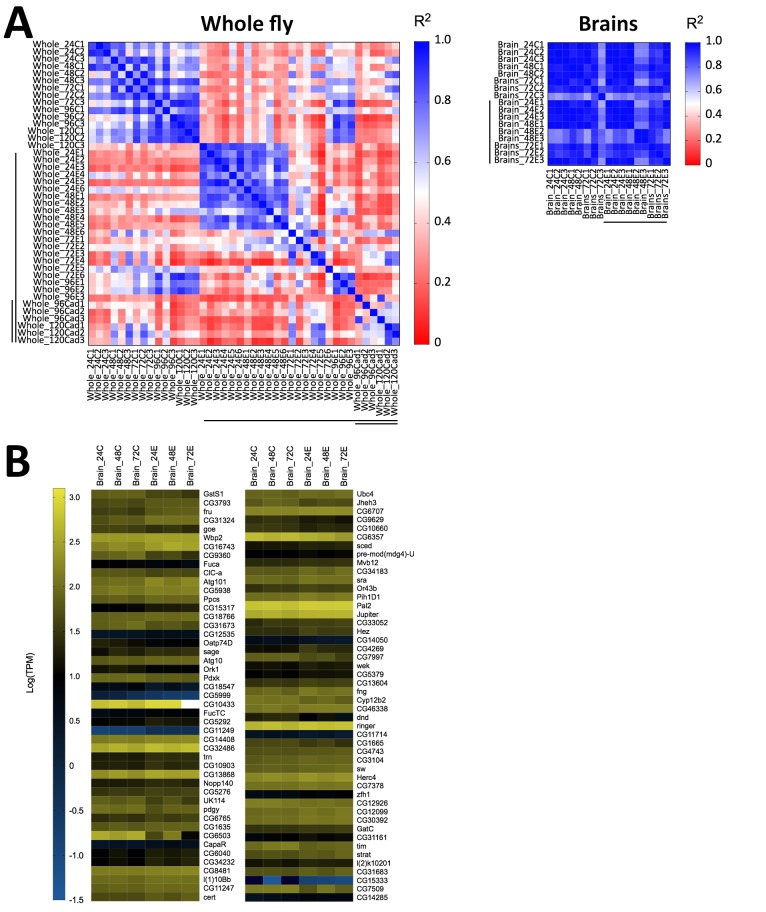
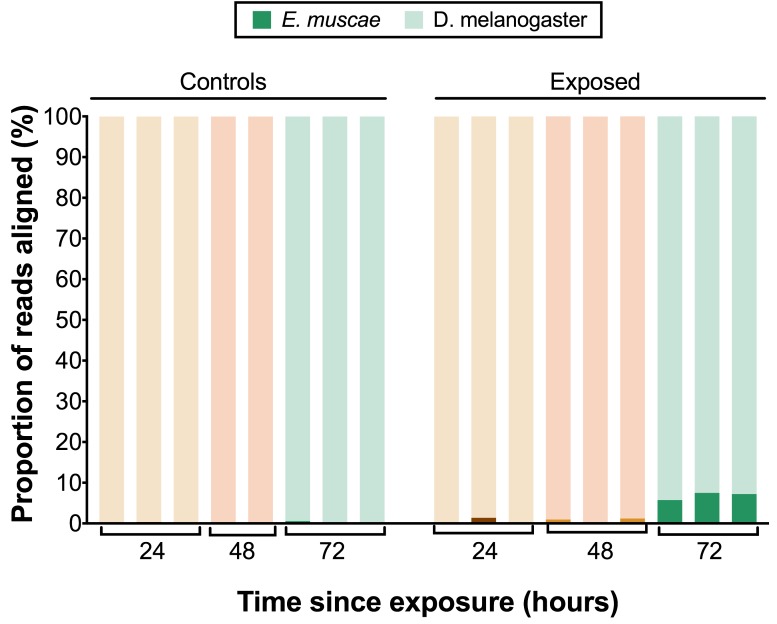
Figure 8. Host gene expression in the brain is minimally altered over the first 72 hr of E. muscae 'Berkeley' infection.
(A) Left) All pairwise linear correlations between samples from E. muscae 'Berkeley'- infected whole fly RNAseq time course. Right) All pairwise linear correlations between samples from E. muscae 'Berkeley'-infected dissected brain pilot RNAseq time course. Samples are named in the following format: Tissue_HourTreatmentReplicate, with ‘C’ indicating controls, ‘E’ indicating exposed flies living at the time sampled and ‘cad’ indicates that the fly had been killed by E. muscae 'Berkeley' before sampling. For example, ‘Whole_24C1’ indicates the first replicate control sample (uninfected fly) of a whole animal taken at 24 hr after mock exposure. Living, exposed samples are denoted on each axis with a black bar; infected cadaver samples are denoted with a double black bar. (B) Heatmap showing all genes that are differentially expressed (p value < 0.001) between controls (uninfected) and exposed flies at 24–72 hr, as determined by ANOVA. Genes are shown in order of increasing p-value from top left corner to bottom right corner.