Figure 1.
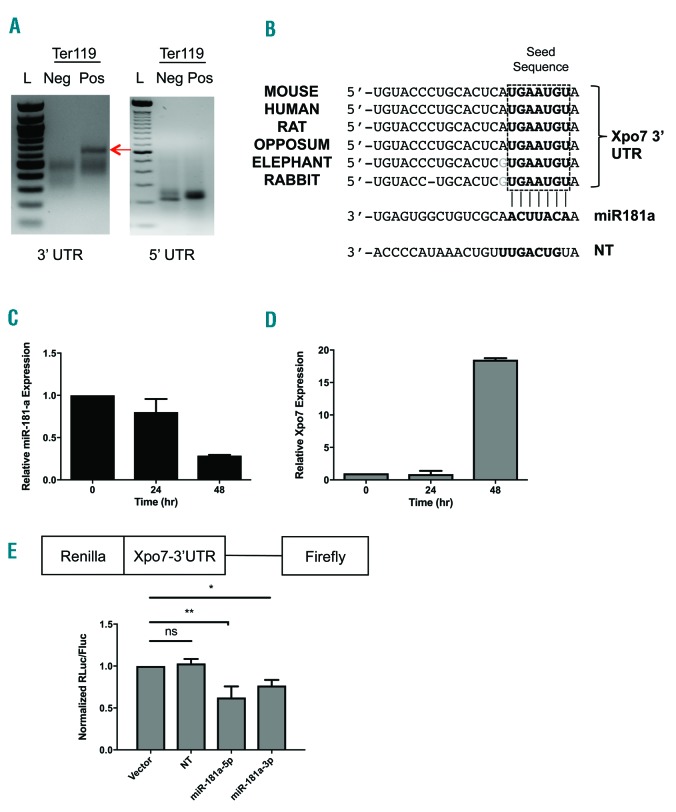
Xpo7 is regulated by erythroid-specific UTRs and is a direct target of miR-181a. (A) 3′- and 5′- RACE (Rapid Amplification of cDNA ends) of RNA isolated from Ter119-negative (non-erythroid) and Ter119-positive (erythroid) cells from murine fetal liver, using freshly isolated total RNA and a RACE kit (Roche). Prominent 3′- and 5′- UTRs change between Ter119-negative (Neg) and Ter119-positive (Pos) cells (representative gels). (B) Significant conservation of a predicted miR-181 site in Xpo7 3′UTR between numerous mammalian species (top). miR-181a complementary seed sequence is shown below that. Seed sequence of the non-targeting (NT) control miRNA (miR-223) is also shown (bottom). (C) qPCR against mature miR-181a transcript was performed using total RNA isolated using a miRNA kit (Qiagen) from erythroblasts during in vitro erythroid differentiation using erythropoietin (Epo) [from time 0–48 hours (hr)]. MiR-181a is developmentally down-regulated during erythroid differentiation. (D) qPCR against Xpo7 shows induction late in the in vitro erythroid differentiation culture. (E) Construct (above) for a luciferase assay using the erythroid (Ter119-positive) Xpo7-3′UTR fused to Renilla luciferase. Fluorescence after co-expression with either miR-181a-5p or -3p mimic was normalized to Firefly luciferase and shows direct binding of miR-181a to the Xpo7-3′UTR. Results are shown as the mean±Standard Error (n=3 per group; *P<0.05, **P<0.01).