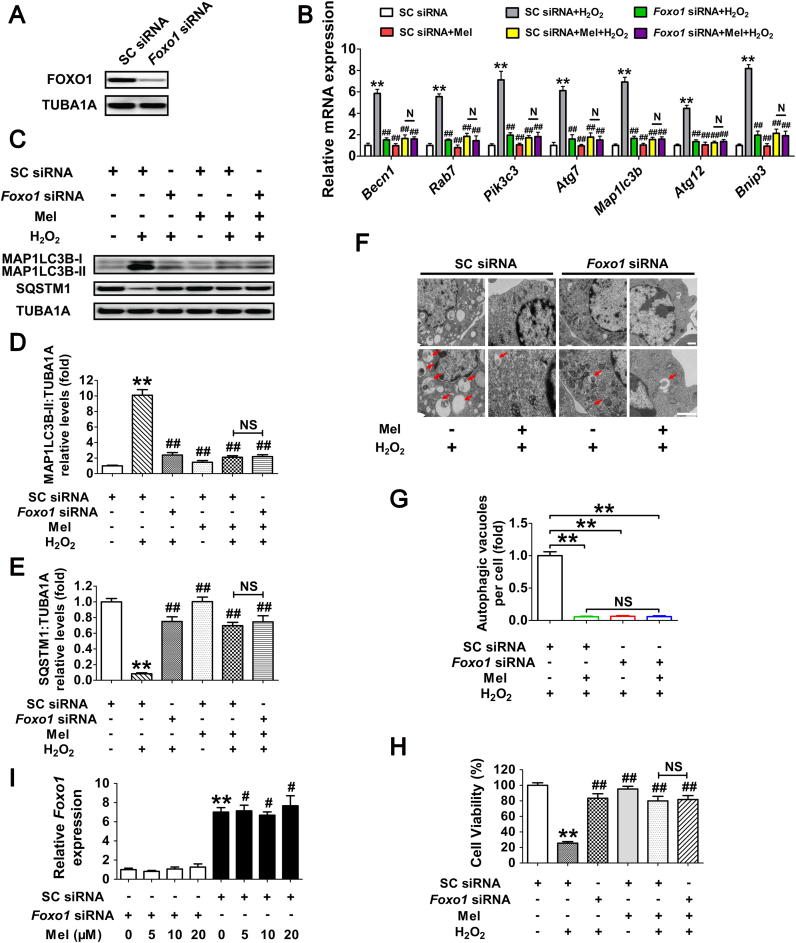
Fig. 6.
Suppression of FOXO1-dependent autophagy by melatonin attenuates oxidative damage in GCs. (A) Primary cultured GCs were transfected with Foxo1 siRNA or scrambled control siRNA for 48 h. The expression of FOXO1 was determined by western blotting. (B) GCs transfected with Foxo1 siRNA or scrambled control siRNA were cultured with 10 μM melatonin for 24 h, washed in PBS, and then subjected to 2 h of H2O2 exposure (200 μM). qRT-PCR was performed to measure the mRNA levels of autophagy-related (Atg) genes in GCs. Expression data were normalized to that of Actb. Significances were marked as **P < 0.01 vs. SC siRNA group; ##P < 0.01 vs. SC siRNA+H2O2 group. N, not significant, P > 0.05. SC siRNA, scrambled control siRNA. (C) Immunoblot analysis of MAP1LC3B and SQSTM1 in GCs with the indicated treatments as described above. (D and E) The MAP1LC3B-II accumulation and SQSTM1 degradation were quantified by densitometric analysis. TUBA1A served as the control for loading. Data represent mean ± S.E; n = 3. ** Represents P < 0.01 vs. control group; ## Represents P < 0.01 vs·H2O2 group. NS, not significant, P > 0.05. (F) GCs transfected with Foxo1 siRNA or scrambled control siRNA for 24 h were cultured for another 24 h in the presence or absence of 10 μM melatonin before 2 h of H2O2 (200 μM) incubation. Cells were then collected for TEM imaging of the autophagic structures. Bar, 1 µm. Enlarged images (below) show clearer autophagic vacuoles (red arrows). (G) Number of autophagic vacuoles per cell section in GCs. Bar graphs are mean ± S.E of results from 10 cell sections. **P < 0.01; NS, not significant, P > 0.05. (H) The detection of cell viability by CCK-8 assay. GCs were treated as above. Data represent mean ± S.E; n = 3 in each group. ** Represents P < 0.01 vs. scrambled siRNA-only-treated cells; ## Represents P < 0.01 vs. H2O2 group. NS, not significant, P > 0.05. (I) GCs transfected with Foxo1 siRNA or scrambled control siRNA for 24 h were exposed to various concentrations of melatonin (0, 5, 10, 20 μM). 24 h later, cells were washed in PBS and incubated with H2O2 (200 μM) for another 2 h. The expression of Foxo1 was determined by qRT-PCR. The relative expression level was normalized to that of Actb. Data represent mean ± S.E; n = 3. ** Represents P < 0.01 vs. Foxo1 siRNA group; # Represents P < 0.01 vs. SC siRNA group. SC siRNA, scrambled control siRNA (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).