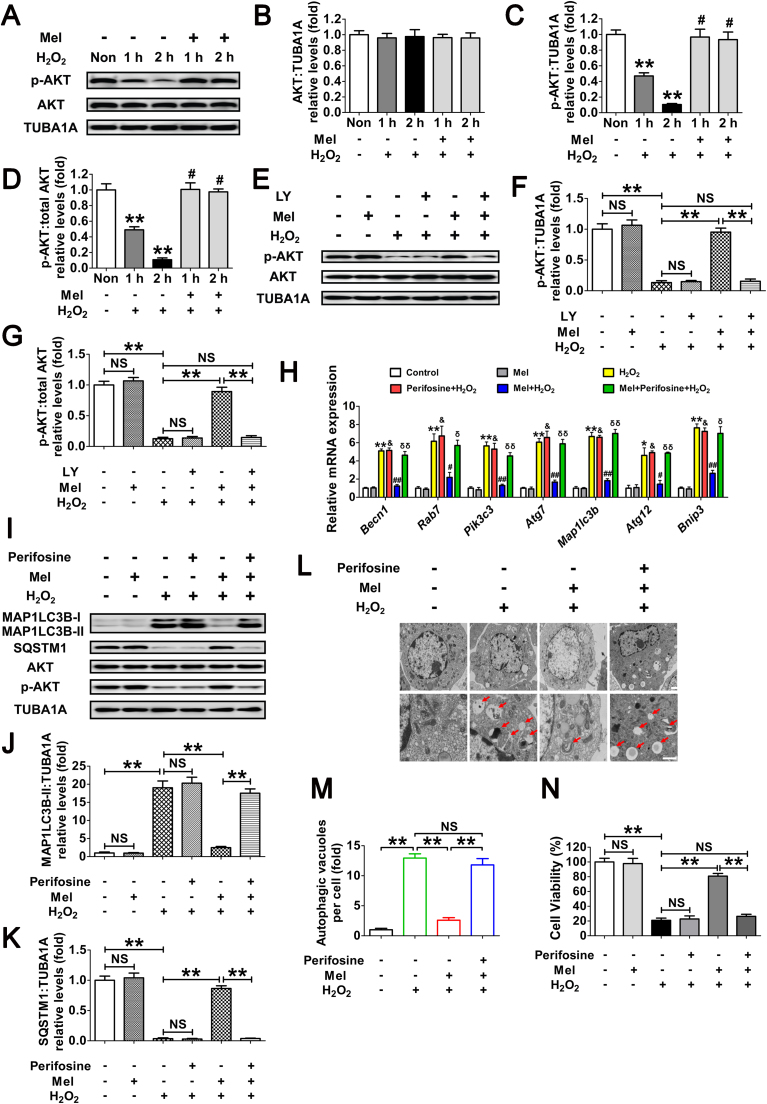
Fig. 7.
Melatonin counteracts H2O2-induced autophagic GC death through the PI3K-AKT pathway. (A) After culturing with 10 μM melatonin for 24 h, GCs were washed in PBS, and then subjected to H2O2 (200 μM) incubation for 1 or 2 h. The expression of phosphorylated AKT (p-AKT) and total AKT was determined by western blotting. (B–D) The relative expression of total AKT, p-AKT, and the ratio of p-AKT to total AKT were quantified using densitometric analysis. TUBA1A served as the control for loading. Data represent mean ± S.E; n = 3. ** Represents P < 0.01 compared to control group. # Represents P > 0.05 compared to control group. (E) GCs grown in medium containing 10 μM melatonin for 24 h were washed in PBS, and then incubated with 200 μM H2O2 for 2 h. For the inhibition of PI3K activity, LY294002 (20 μM) was added 1 h before H2O2 treatment. Western blotting was performed to measure the protein levels of p-AKT and total AKT. (F and G) Quantification of p-AKT expression and the ratio of p-AKT to total AKT. **P < 0.01; NS, not significant, P > 0.05. (H) GCs pretreated with or without 10 μM melatonin for 24 h were then rinsed in PBS, and cultured in the presence or absence of 200 μM H2O2 for 2 h. qRT-PCR was performed to measure the mRNA levels of autophagy-related (Atg) genes in GCs. Expression data were normalized to that of Actb. Data represent mean ± S.E; n = 3 in each group. * Represents P < 0.05 (** Represents P < 0.01) compared to control group. & Represents P > 0.05 compared to H2O2-only-treated cells. # Represents P < 0.05 (## Represents P < 0.01) compared to H2O2-only-treated cells. δ Represents P < 0.05 (δδ Represents P < 0.01) compared to melatonin+H2O2 group. (I) Immunoblotting analysis of MAP1LC3B, SQSTM1, AKT and p-AKT in GCs with the indicated treatments as described above. (J and K) Quantification of the MAP1LC3B-II accumulation and SQSTM1 degradation. Data represent mean ± S.E; n = 3. **P < 0.01; NS, not significant, P > 0.05. (L) TEM imaging of the autophagic structures in GCs with indicated treatments as mentioned above. Bar, 1 µm. Enlarged images (below) show clearer autophagic vacuoles (red arrows). (M) Number of autophagic vacuoles per cell section in GCs. Bar graphs are mean ± S.E of results from 10 cell sections. **P < 0.01; NS, not significant, P > 0.05. (N) Cell viability was determined by CCK-8 assay in GCs received the indicated treatments. Data represent mean ± S.E; n = 3. **P < 0.01; NS, not significant, P > 0.05 (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).