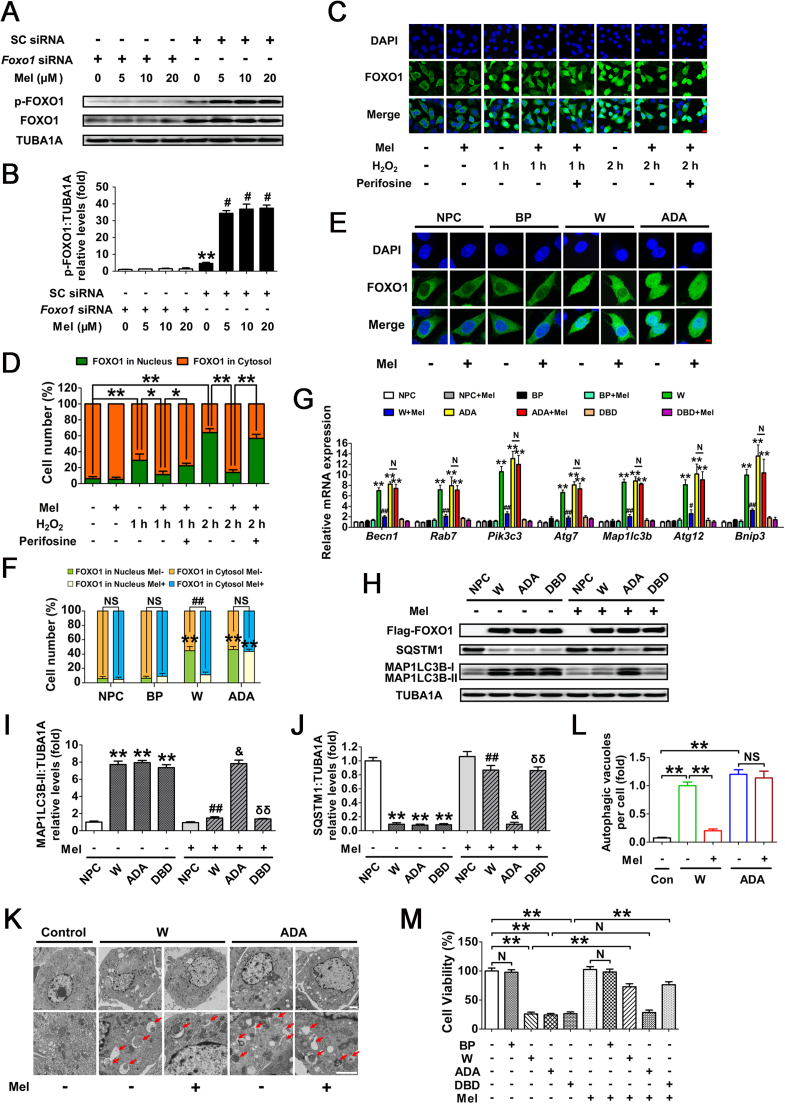
Fig. 8.
Inhibition of FOXO1 transcriptional activity through the melatonin-PI3K-AKT axis protects GCs from H2O2-induced autophagic PCD. (A) GCs transfected with Foxo1 siRNA or scrambled control siRNA for 24 h were cultured in media containing various concentrations of melatonin (0, 5, 10, 20 μM). 24 h later, cells were rinsed with PBS, and exposed to H2O2 (200 μM) incubation for another 2 h. The expression of phosphorylated FOXO1 (p-FOXO1) was determined by western blotting. (B) The phosphorylation level of FOXO1 was quantified by densitometric analysis. TUBA1A served as the control for loading. Data represent mean ± S.E; n = 3. ** Represents P < 0.01 vs. Foxo1 siRNA group; # Represents P < 0.01 vs. SC siRNA group. SC siRNA, scrambled control siRNA. (C) GCs pretreated with or without 10 μM melatonin for 24 h were then rinsed in PBS, and subjected to 1 or 2 h of H2O2 (200 μM) incubation. For the inhibition of AKT, Perifosine (10 μM) was added 1 h before H2O2 exposure. Subcellular localization of FOXO1 was detected using anti-FOXO1 (green), and the nuclei were counterstained with DAPI (blue). Bar, 10 µm. (D) The percentage of cells with FOXO1 in the nucleus (green bars) and in the cytosol (orange bars). Experiments were repeated in triplicate, and 3 fields of each coverslip were selected in random for counting. Data represent mean ± S.E; n = 3. *P < 0.05, **P < 0.01. (E) A Flag-tagged FOXO1 (WT), FOXO1T24A,S253D,S316A (ADA), or an empty control plasmid (BP) was individually transfected into GCs. 24 h later, cells were cultured with or without melatonin (10 μM) for another 24 h. Immunofluorescence microscopy was performed to visualize subcellular localization of FOXO1 (green). The nuclei were counterstained with DAPI (blue). Bar, 5 µm. (F) The percentage of cells with FOXO1 in the nucleus or the cytosol under the indicated treatments. NPC, non-plasmid control; BP, blank plasmid; W, FOXO1-WT plasmid; ADA, FOXO1T24A,S253D,S316A plasmid. ** Represents P < 0.01 compared with the non-plasmid control; NS, not significant, P > 0.05; ##, P < 0.01. (G) qRT-PCR analysis of Atg genes transcription in GCs transfected with FOXO1-expressing vectors (FOXO1-WT, FOXO1T24A,S253D,S316A, and FOXO1N208A,H212R) in the presence or absence of melatonin (10 μM) as mentioned above. Expression data were normalized to that of Actb. Data represent mean ± S.E; n = 3 in each group. ** Represents P < 0.01 compared with nonplasmid control. # Represents P < 0.05 (## Represents P < 0.01) compared with FOXO1-WT (W) group. N Represents P > 0.05 compared with the FOXO1T24A,S253D,S316A (ADA) group. (H) Western blotting was performed to measure the protein levels of Flag-FOXO1, MAP1LC3B and SQSTM1 in GCs with the indicated treatments as described above. (I and J) Quantification of MAP1LC3B-II accumulation and SQSTM1 degradation. TUBA1A served as the control for loading. *P < 0.05 (**P < 0.01) vs. the non-plasmid control without melatonin treatment. ## Represents P < 0.01 vs. FOXO1-WT-transfected group without melatonin treatment. & Represents P > 0.05 vs. FOXO1T24A,S253D,S316A (ADA)-transfected group without melatonin treatment. δδ Represents P < 0.01 vs. FOXO1N208A,H212R (DBD)-transfected group without melatonin treatment. (K) Primary cultured GCs remained as an untreated control or were transfected with FOXO1-WT or FOXO1T24A,S253D,S316A for 24 h. Cells were then grown for another 24 h in the presence or absence of 10 μM melatonin, and collected for TEM imaging of the autophagic structures. Bar, 1 µm. Enlarged images (below) show clearer autophagic vacuoles (red arrows). (L) Number of autophagic vacuoles per cell section in GCs. Bar graphs are mean ± S.E of results from 10 cell sections. **P < 0.01; NS, not significant, P > 0.05. (M) Cell viability was measured by CCK-8 assay in GCs transfected with FOXO1 expression plasmids upon melatonin (10 μM) treatment. **P < 0.01; N, not significant, P > 0.05. BP, blank plasmid.