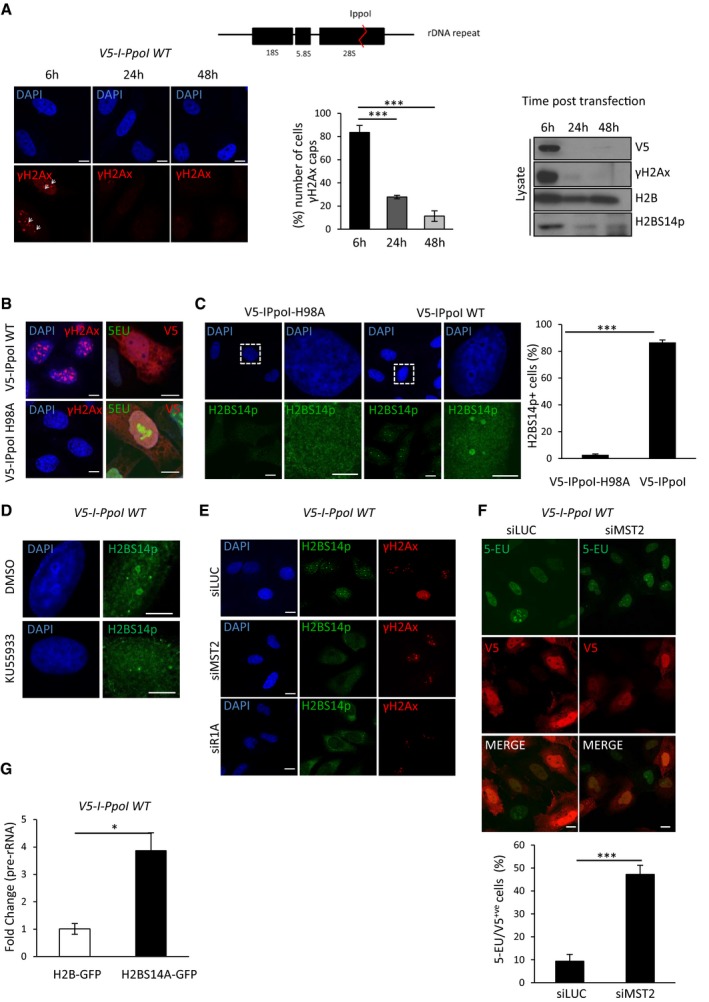
I‐PpoI recognises a 15 bp sequence in the rDNA repeat (top); In vitro transcribed mRNA of a V5 epitope‐tagged derivative was directly transfected into HeLa cells. Cells were lysed at the indicated times and analysed with Western blot for the indicated antibodies (lower right). rDNA repair was measured by the presence of γH2AX. Representative images (lower left) and quantification (lower middle) of cells with γH2AX‐positive nucleolar caps are shown. Arrowheads point at γH2AX‐positive nucleolar caps. Error bars represent SD and derive from three independent experiments.
HeLa cells were transfected with mRNA from V5‐I‐PpoI WT or catalytically inactive I‐PpoI H98A. 6 h post‐mRNA transfection accumulation of γH2AX and 5‐EU incorporation was assessed.
I‐PpoI WT or I‐PpoI H98A mRNA was transfected in HeLa cells, 6 h post‐transfection cells was fixed and stained for H2BS14p. Boxed areas are shown in higher magnification. Representative images and quantification of H2BS14p‐positive cells are shown. Error bars represent the SD and derive from three independent experiments.
HeLa cells were treated or not with 10 μM ATM inhibitor (KU55933), transfected with I‐PpoI WT mRNA as above, followed by fixation and staining for H2BS14p.
HeLa cells were initially transfected with the indicated siRNAs and I‐PpoI WT mRNA introduced after 48 h, cells were stained for the indicated antibodies.
HeLa cells were initially transfected with siMST2 or control siRNA and I‐PpoI WT mRNA introduced after 48 h. Six hours post‐mRNA transfection cells were assessed for I‐PpoI expression and 5‐EU incorporation. Quantifications and representative images are shown. Error bars represent the SD and derive from three independent experiments.
HeLa cells were transfected with H2B‐GFP or H2BS14A‐GFP. rDNA DSBs were introduced transfecting by I‐PpoI‐WT mRNA. Pre‐rRNA expression relative to GAPDH was assessed with qPCR. Error bars represent the SD and derive from two independent experiments.
Data information: Scale bars 10 μm. Two‐tailed Student's
‐test was used for statistical analysis. *
< 0.001.