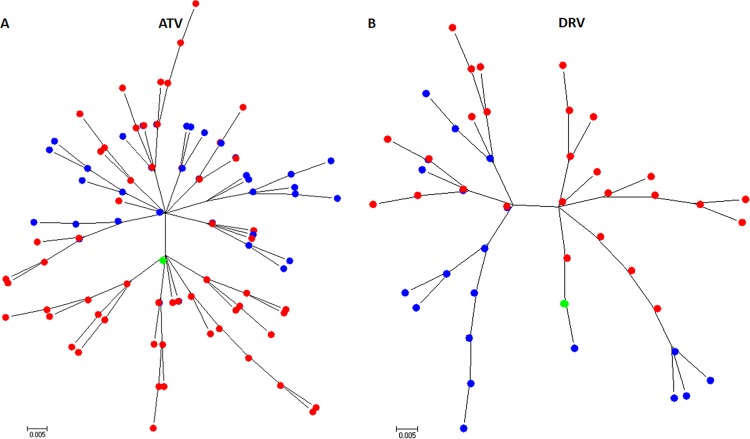
FIG 3.
Maximum likelihood phylogram of wild-type (WT) and MAX unique HIV-1 protease amino acid variants selected after 32 passages in MT-4 cells and in the presence of atazanavir (ATV) (A) or darunavir (DRV) (B). Phylogenetic reconstruction was generated using a Jones-Taylor Thornton (JTT) model as implemented in the MEGA6 software package. Both phylogenetic trees show that the WT and MAX viruses, which shared an identical starting amino acid sequence, followed different evolutionary trajectories. Blue and red labels correspond to WT and MAX variants, respectively. Green labels represent the starting HXB2 protease amino acid sequence.