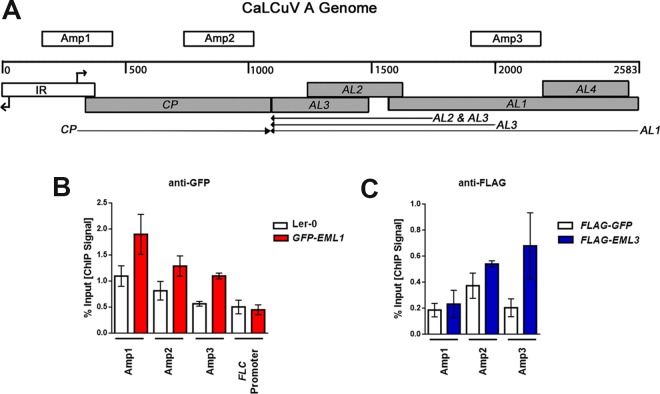
FIG 3.
EML1 and EML3 associate with similar regions of CaLCuV DNA A. (A) Linear representation of the circular dsDNA A genome component (2,583 bp), showing viral open reading frames (shaded boxes), the intergenic region (IR) containing divergent promoters and transcription start sites (right-angle arrows), viral transcripts (arrows), and the amplicons (Amp 1, 2, and 3) tested in ChIP experiments. (B) ChIP-qPCR was performed with nuclear extracts from silique and floral tissues of CaLCuV-infected GFP-EML1 transgenic plants and Ler-0 control plants, using GFP antibody and primers amplifying the indicated amplicons. An amplicon within the FLC promoter served as a negative control. (C) ChIP-qPCR was performed with nuclear extracts from N. benthamiana leaf tissue coinfiltrated with constructs to express CaLCuV DNA A, CaLCuV DNA B, and FLAG-EML3 or FLAG-GFP (control), using FLAG antibody and primers amplifying the indicated amplicons. Bars indicate standard errors for data compiled from at least three biological replicates, each with at least two technical replicates.