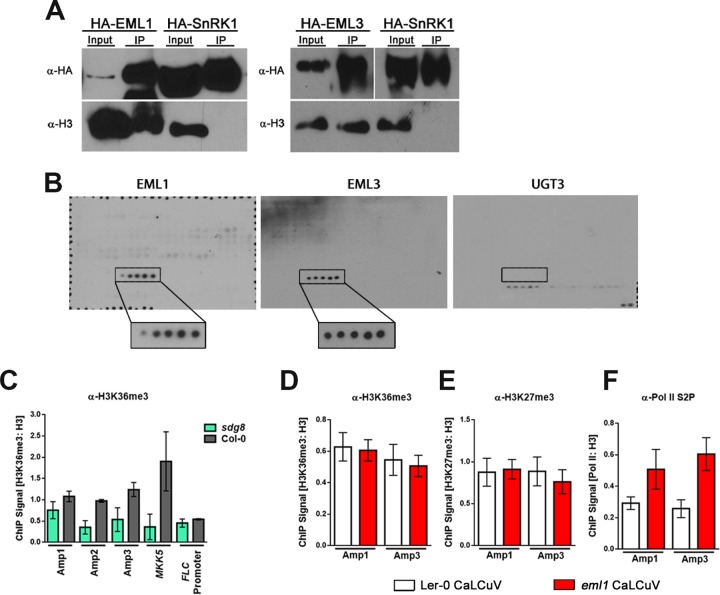
FIG 4.
EML1 and EML3 are histone readers that recognize H3K36. (A) HA-tagged EML proteins and the kinase domain of SnRK1 (control) were transiently expressed in N. benthamiana leaves. Immunoprecipitation (IP) was performed with HA antibody (α-HA), and immunodetection was performed with HA antibody or histone H3 antibody (α-H3) as a proxy for nucleosomes. (B) Purified His-tagged EML1, EML3, and UGT3 (control) protein association with the MODified histone peptide array (Active Motif). Positive signals detected with His-EML1 and His-EML3 are boxed, with boxes enlarged for increased visibility. Positive signals represent H3 peptide fragments (amino acids 26 to 45) containing, from left to right, unmodified H3K36, H3K36me1, H3K36me2, H3K36me3, and H3K36ac. The box on the UGT3 blot marks the position of the same H3K36-containing peptide fragments for which no binding was detected. Detected signals on this blot are a result of nonspecific binding, based on the presence of signals representing background control peptides (bottom right) after a 3-fold-longer exposure. Probing was performed 3 times for each protein, and the representative blots are shown. (C) ChIP-qPCR was performed with nuclear extracts from silique and floral tissues of wild-type (Col-0) or sdg8 plants infected with CaLCuV using H3K36me3 antibody (α-H3K36me3) and primers amplifying the indicated regions of CaLCuV DNA A (Fig. 3A) or positive-control (MKK5) and negative-control (FLC promoter) loci. Values were normalized to data from ChIP-qPCRs performed with histone H3 antibody using the same extracts. (D to F) ChIP-qPCR was performed with nuclear extracts from silique and floral tissues of Ler-0 (wild-type) and eml1 plants infected with CaLCuV, using H3K36me3 antibody (D), H3K27me3 antibody (E), or elongating Pol II antibody (Pol II phosphorylated at C-terminal domain serine 2 [Pol II S2P]) (F). Primers employed amplified the indicated regions of CaLCuV DNA A. Values were normalized to data from ChIP-qPCRs performed with H3 antibody using the same extracts. Bars indicate standard errors for a minimum of three biological replicates, each with at least two technical replicates.