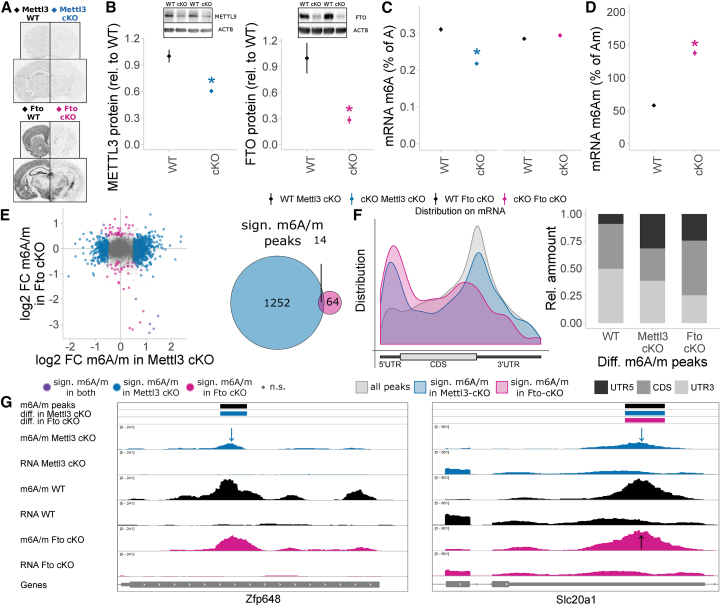
Figure 4.
Depletion of METTL3 and FTO in Adult Excitatory Neurons Using the Camk2a-Cre Driver Changes the Cortex Epitranscriptome
(A) Mettl3 and Fto mRNA are depleted from the neocortex and hippocampus in Mettl3 cKO and Fto cKO mice, respectively. In situ hybridization, n = 3, representative shown. WT, wild-type; cKO, conditional.
(B) METTL3 and FTO proteins are significantly depleted in Mettl3 cKO and Fto cKO mice, respectively. Western blot of PFC protein, optical density normalized from digitally acquired image signal normalized to ACTB protein. n = 4–5, mean ± SEM. ∗p < 0.05, t test. For full blots, see Figure S5.
(C) Global mRNA m6A is decreased in Mettl3 cKO mice, but not in Fto cKO mice, when measured with LC-MS/MS. n = 5, mean ± SEM, m6A-specific measurement. Two-way ANOVA interaction effect F(1, 19) = 106.269, p < 0.001. ∗p < 0.05, omnibus Tukey post hoc tests to respective WT. See also Figure S5.
(D) Global mRNA m6Am is increased in Fto cKO mice when measured with LC-MS/MS. n = 5, mean ± SEM, m6Am-specific measurement. Data are shown relative to Am, which is not altered in Fto cKO mice. ∗p < 0.05, t test. For LC-MS/MS traces, see Figure S5.
(E) The m6A/m epitranscriptome is widely altered in in Mettl3 cKO and Fto cKO mice. m6A/m-seq on mouse cortex poly(A)-RNA of WT and cKO animals reported 1,266 and 78 significantly different methylated m6A/m peaks in Mettl3 cKO and in Fto cKO compared to WT, respectively, with 14 shared peaks. n = 3–5, each pooled from 3 mice. WT of both lines were grouped together as we observed no major regulation between them. Shown are log2 fold changes of methylation in cKO relative to WT mice using 10,109 high-confidence consensus m6A/m peaks detected across all groups, mapping to 6,056 unique genes. Significantly regulated m6A/m peaks are Q < 0.1 and absolute log2 fold change > 0.5.
(F) m6A/m peaks are enriched at the stop codon with a less prominent enrichment at the 5′ UTR, as observed in Figure 1. Differentially methylated peaks in both Mettl3 cKO and Fto cKO mice show an increased preference for 5′ UTR position with a decreased preference for CDS peaks in Mettl3 cKO differential peaks. Peak distribution mapped along mRNA relative position.
(G) Two examples m6A/m peaks regulated only in Mettl3 cKO or in both Mettl3 cKO and Fto cKO. Shown are averaged sequence tracks m6A/m-seq and RNA-seq per group and detected m6A/m peaks. Arrows indicate quantitatively regulated peaks (Q < 0.1, absolute log2 fold change >0.5).