Figure 7.
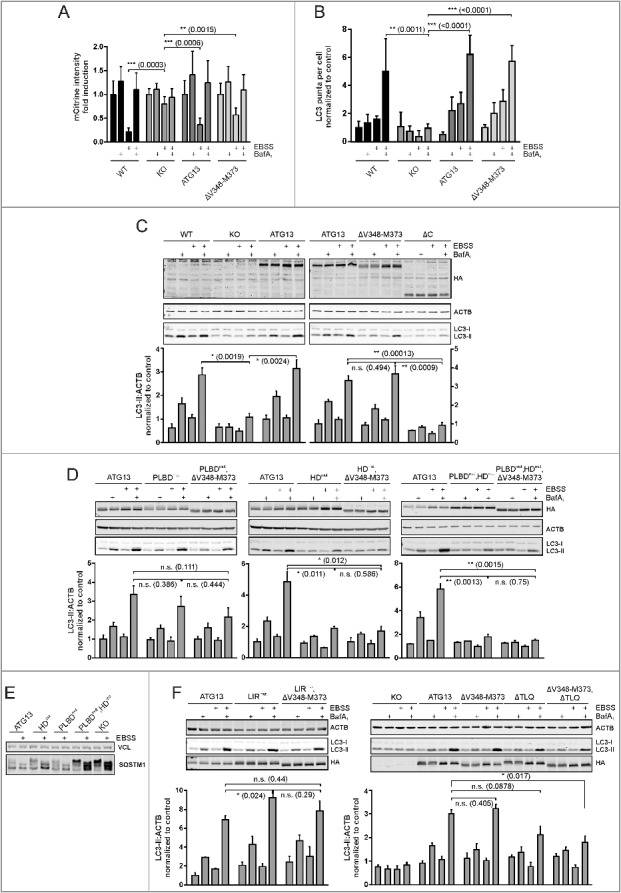
Differential requirement of ATG13 interaction interfaces for amino acid starvation-induced LC3 turnover. (A) Atg13 wild-type MEFs (WT) or atg13 KO MEFs stably expressing mCitrine-LC3B and the indicated ATG13 variants were cultured in growth medium or starvation medium (EBSS) with or without 40 nM bafilomycin A1 (BafA1) for 8 h. Total cellular mCitrine-LC3B signals were analyzed by flow cytometry. The median of fluorescence intensity for each sample was normalized to wild-type cells incubated in growth medium. Data represent mean + SEM. **P < 0.01, ***P < 0.001 (Student t test, 2-sample assuming unequal variances). (B) Untransfected Atg13 wild-type (WT) MEFs or atg13 KO MEFs retrovirally transfected with empty vector (KO) or cDNA encoding the indicated ATG13 variants were grown on glass cover slips overnight and incubated for 2 h in growth medium or starvation medium (EBSS) in the presence or absence of 40 nM bafilomycin A1 (BafA1). Cells were fixed, permeabilized and stained for LC3. Imaging was performed using an inverse confocal laser scanning microscope and puncta per cell quantification was done using fiji software. Data represent mean + SEM. Statistical analysis using the Student t test, 2-sample assuming unequal variances was performed comparing LC3 puncta accumulation during EBSS + BafA1 treatment for depicted cell lines. **P < 0.01, ***P < 0.001 (Student t test, 2-sample assuming unequal variances). (C, D, F) Untransfected Atg13 wild-type MEFs (WT) or atg13 KO MEFs retrovirally transfected with empty vector (KO) or cDNA encoding the indicated ATG13 variants were incubated as described in (B). Cleared cellular lysates were analyzed by immunoblotting for HA, LC3, ACTB, and VCL. Fold changes were calculated by dividing each normalized ratio (protein to loading control) by the average of the ratios of the control lane (ATG13 in medium). Results are mean + SEM *P < 0.05, **P < 0.01, n.s., not significant (Student t test, 2-sample assuming unequal variances). (E) atg13 KO MEFs retrovirally transfected with empty vector (KO) or cDNA encoding the indicated ATG13 variants were incubated in growth medium or starvation medium (EBSS) for 2 h. Cleared cellular lysates were analyzed by immunoblotting for SQSTM1 or VCL.
