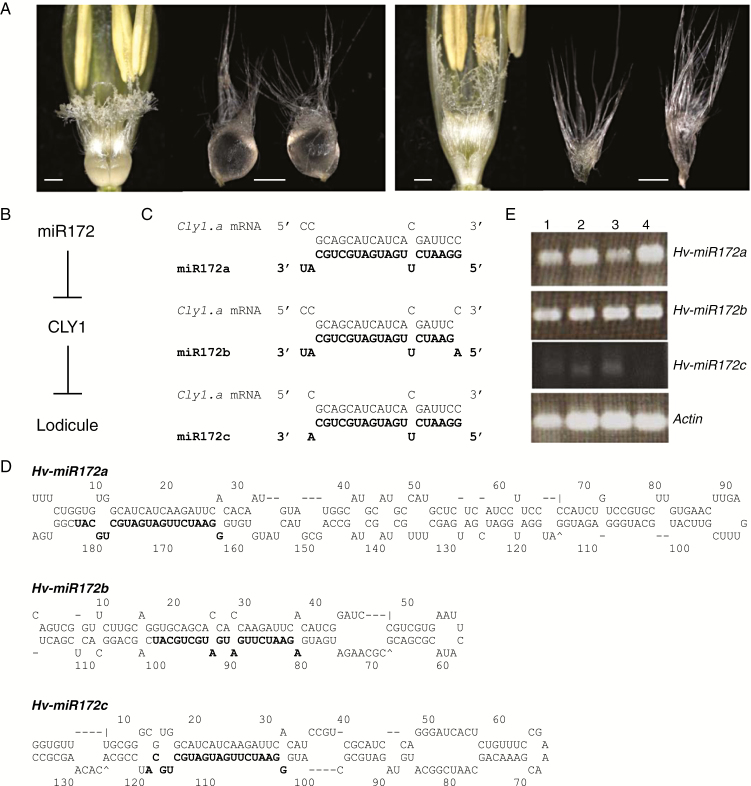
Fig. 1.
The barley inflorescence and the Hv-miR172 genes. (A) Variation in lodicule size at anthesis. (Left) AZ (non-cleistogamous). (Right) KNG (cleistogamous). Scale bar = 0.5 mm. (B) Scheme of proposed interaction between miR172 and CLY1 for lodicule development. The Cly1.a and cly1.b alleles both encode CLY1, which suppresses the development of lodicules. Expression of Cly1.a is negatively regulated by miR172, resulting in less suppression of lodicule development by CLY1. (C) Potential interactions between cly1 mRNA and miR172 isoforms in cultivar ‘Morex’. Bold letters indicate miR172 sequences that can potentially bind with cly1 mRNA sequence. (D) In silico predicted hairpin secondary structure of the miR172 precursors. Bold text indicates the mature miRNA sequence. Precursor sequence length and free folding energy (ΔG) for Hv-miR172a, b and c were, respectively, 189 nt and −92.80 kcal mol−1, 117 nt and −58.20 kcal mol−1 and 135 nt and −63.60 kcal mol−1. (E) RT–PCR-derived abundance of primary miR172s in the immature spikes at the awn primordium stage. Lane 1, AZ; lane 2, KNG; lane 3, ‘Morex’; lane 4, ‘Golden Promise’. Actin was the chosen reference sequence.