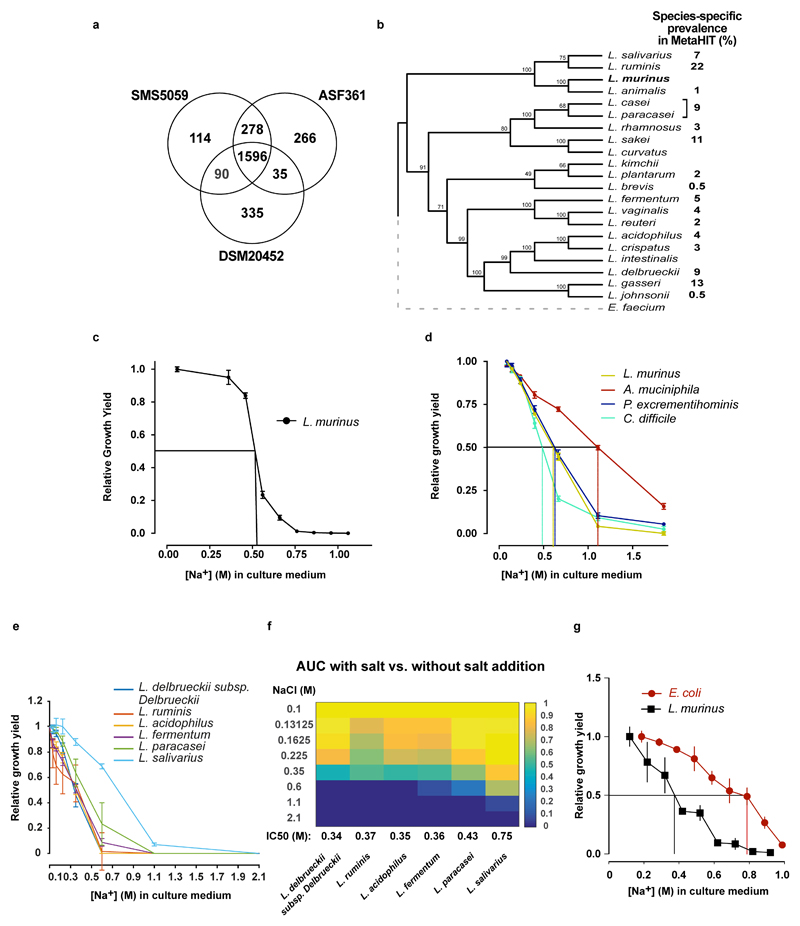
Extended Data Figure 5. L. murinus genome and in vitro growth of Lactobacilli.
(a) Venn diagram of the coding sequences present in the L. murinus and two other isolates with available with full genome sequences. (b) Bootstrapped phylogenetic tree of full-length 16S rDNA from a variety of Lactobacillus species resident to rodent or human guts. Prevalence of the respective species in the MetaHIT cohort is shown. L. murinus strains are absent in the MetaHIT cohort. (c) Growth yield (OD600) of L. murinus measured at increasing concentrations of NaCl. Aerobic endpoint measurements of liquid L. murinus cultures in MRS medium and increasing NaCl concentrations relative to growth in MRS without the addition of NaCl. n=5 independent experiments. (d) Anaerobic growth yield of L. murinus, A. muciniphila, P. excrementihominis, and C. difficile grown at 37 °C for 48 hours in MGAM liquid medium. Growth at each salt concentration is normalized to growth at 0.086 M Na+. The respective IC50 is indicated. n=3 technical replicates across 2 experiments. (e) Anaerobic growth of selected human Lactobacilli in MGAM medium with increasing NaCl concentrations. Relative growth yield is calculated based on AUCs by comparing to growth in MGAM without the addition of NaCl. n=3 independent experiments with 3 technical replicates. (f) Heatmap showing data as in (e). The respective IC50 is shown in the bottom row. (g) Growth yield of E. coli and L. murinus, grown at 37 °C for 12-16 hours on LB (E.coli) or MRS broth (L. murinus). n=4 technical replicates from two independent experiments. Mean±s.e.m.