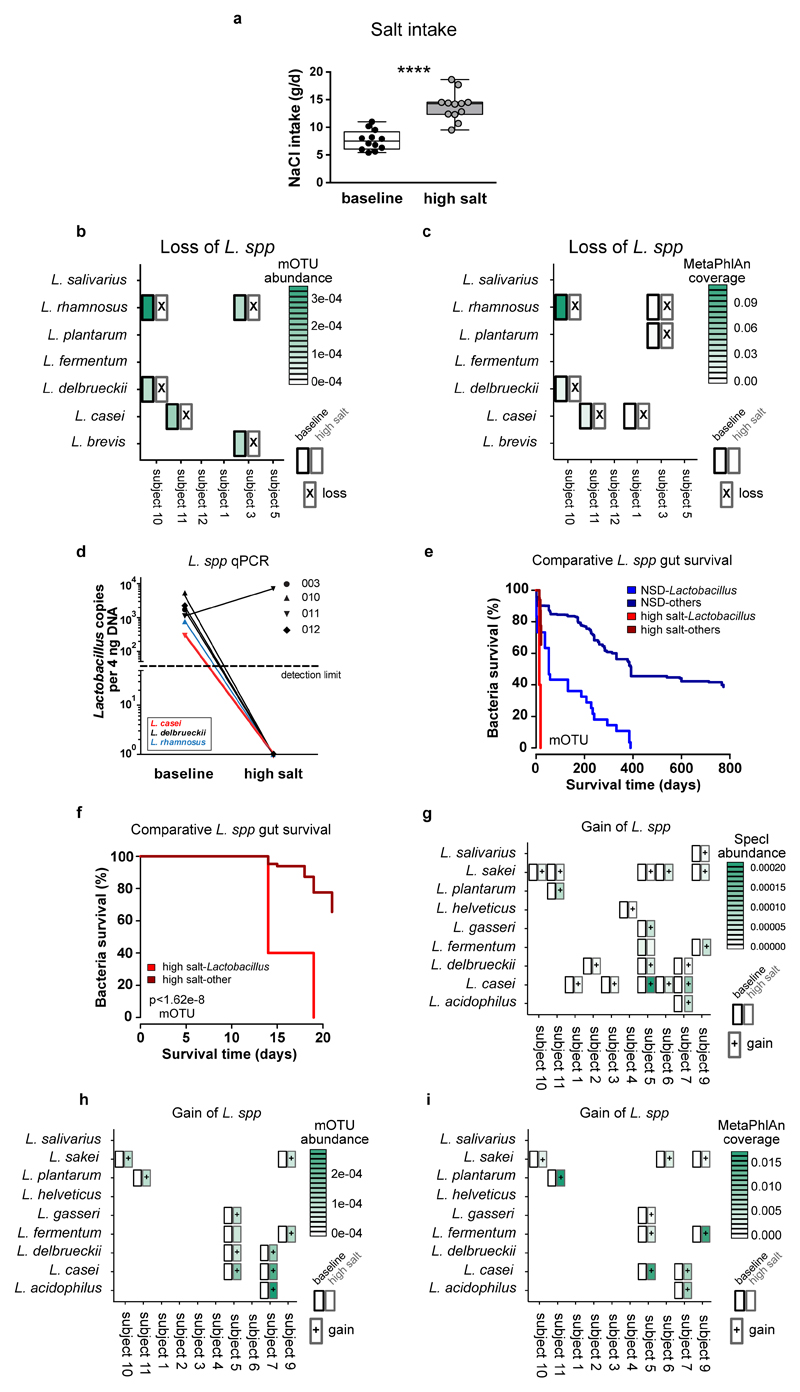
Extended Data Figure 10. High salt challenge in healthy human subjects.
(a) Total salt intake according to dietary records (n=12, paired one-tailed t-test). (b, c) Metagenome analysis shows loss of Lactobacillus gut populations during human high salt challenge. Shown are all subjects (horizontal axis) for which gut Lactobacilli were detected at baseline and all species so detected (vertical axis) using the mOTU (b) or MetaPhlAn framework (c) for bacterial species identification. Heatmap cells show abundance for mOTU (insert counts as fraction of sample total) or average coverage (reads per position) for MetaPhlAn of these Lactobacilli at baseline (left part of cells, black border) and after high salt challenge (right part of cells, grey border). Cross markers show complete loss (nondetection after high salt challenge) of each species. In all cases but one (shown), baseline Lactobacillus populations are no longer detected post high salt. (d) qPCR using Lactobacillus-specific 16S rDNA primers in human fecal samples positive for Lactobacillus at baseline show a loss of the respective species after 14 days of high salt. Lactobacillus 16S rDNA copy number in 4 ng fecal DNA is shown. Symbols indicate study subject, colors indicate respective Lactobacillus species. (e) Kaplan-Meier survival curves contrasting the fate of gut Lactobacillus populations (detected using the mOTU framework) following high salt challenge (bright red curve) and in healthy control individuals from reference cohorts (n=121, see methods) not undergoing any intervention (bright blue curve). This is compared with corresponding survival curves over time for the set of all other detected gut bacterial species following high salt challenge (high salt-others, dark red curve) and without such challenge in controls (NSD-others, dark blue curve). (f) For a clearer view of its time range only the salt intervention curves from (e) are shown. Two observations are clear. First, Lactobacillus on average persist for shorter times in the gut than the average over all other species. Second, a high salt challenge strongly increases gut loss of both Lactobacillus and non-Lactobacillus species. As such, in combination, Lactobacillus loss is highly pronounced under high salt intervention and significantly (p<1.62e-8) faster than the average over all species. (g-i) Metagenome analysis shows introduction of novel Lactobacillus gut populations during human high salt challenge. Shown are all subjects (horizontal axis) for which gut Lactobacilli were detected following high salt challenge, and all species so detected (vertical axis) using the SpecI (g), mOTU (h) or MetaPhlAn (i). Heatmap cells show abundance (insert counts as fraction of sample total for SpecI and mOTU) and average coverage (reads per position for MetaPhlAn) of these Lactobacilli at baseline (left part of cells, black border) and after high salt challenge (right part of cells, gray border). Cross markers show novel introduction (nondetection at baseline) of each species.