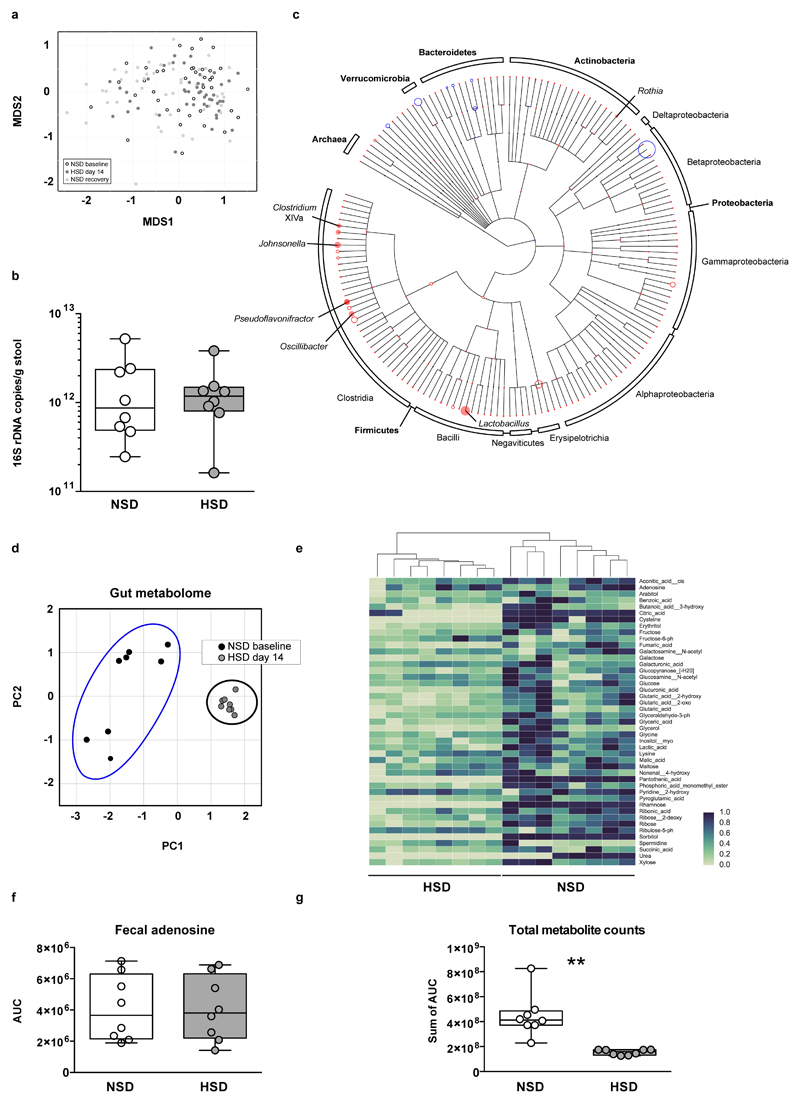
Extended Data Figure 3. HSD alters the fecal microbiome and the fecal metabolite profile.
(a) Mouse 16S rDNA fecal microbiome samples do not separate by diet in a MDS ordination (white, NSD samples; black, HSD samples; grey, recovery on NSD). (b) Real-time PCR on DNA extracted from fecal samples of mice fed NSD or HSD using universal 16S rDNA primers (n=8 fecal samples per group from independent mice, indicated by circles; two-tailed Wilcoxon matched-pairs signed rank test). (c) Phylogenetic tree showing changes in microbiome composition caused by HSD. OTUs present in samples from day 14 are indicated by colored circles (red indicates reduction in HSD samples; blue indicates enrichment). The circles’ radii indicate median log fold difference in relative abundance between the two diets. Filled circles mark statistically significant differences (two-tailed Student‘s t-test, Benjamini-Hochberg correction, p<0.05). (d-g) High dietary salt strongly influences the fecal metabolite profile. Male FVB/N mice (n=8) were fed a NSD and then switched to HSD. Metabolites were extracted from fecal pellets taken under NSD (day -3) and HSD (day 13), and analyzed by gas-chromatography mass-spectrometry. (d) HSD samples are clearly distinguishable from NSD samples in a principal component analysis for fecal metabolites. (e) Fecal metabolites clearly cluster by treatment. The majority of fecal metabolites are reduced by HSD. Hierarchically clustered heatmap, metabolites shown in alphabetical order. Metabolites were normalized by subtracting the minimum and dividing by the maximum value across all mice. (f) Fecal levels of the nucleoside adenosine were similar in both diets, suggesting that the change in metabolites is not due to a decrease in overall bacterial biomass. (g) HSD leads to a reduction in total metabolite peak intensities in fecal samples. **p<0.01 two-tailed paired Student‘s t-test for (f, g).