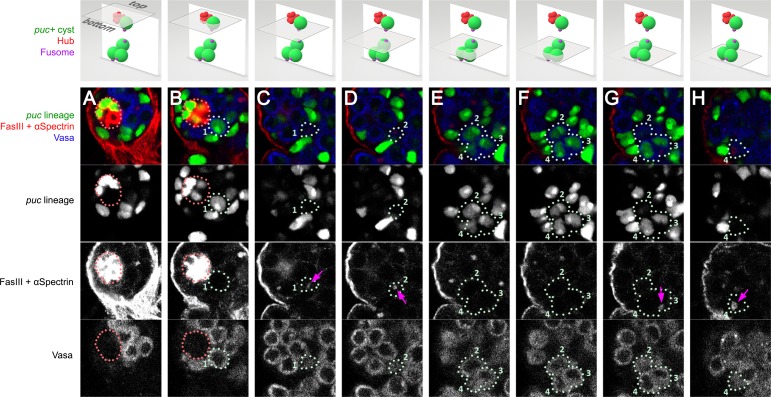
Figure 6. JNK pathway activity is required for bam-lineage spermatogonial dedifferentiation.
(A–E) Representative images of testes where puckeredE69 (puc)-Gal4 was lineage-traced at 0 days fed (A), 15 days fed (B), 20 days fed (C), after 15 days starved (D) and after 15 days starved and 5 days refed (E). Under fed conditions, puc-lineage cells were restricted to some niche cells (asterisk) (A–C). After 15 days of starvation, most testes had puc-lineage labeling of somatic cells (quantified in F) (D). Only during the refeeding phase were puc-lineage germline cells observed (quantified in F) (E). Arrows in E mark puc-lineage-positive GSCs and the bracket in E marks puc-lineage-positive spermatogonia. (F, F’) Graphs indicating the percentage of testes that had pucE69-Gal4- (F) or puc-IT.Gal4- (F’) lineage positive somatic cells (light gray bars) or germline cells including GSCs (dark gray bars) at the time points indicated in A-E, as well as at 2 and 3 days of refeeding in (F). (G–J) Representative images of the testis stem cell niche in control bam-Gal4, UAS-LacZ (bam > LacZ, G, H) or bam-Gal4, UAS-puc (bam > puc, I, J), where we blocked JNK signaling, at 0 days (G,I) of after 15 days of starvation and 5 days of refeeding (H,J). The bam lineage is labeled in green, the germline (Vasa) is red and the niche (FasIII) is blue. GSCs are outlined by the dashed line. There are no GSCs derived from the bam-lineage cells in the control or bam > puc testes at 0 days (G, I). After starvation and refeeding, the control (bam > LacZ) contains GSCs positive for the bam-lineage (indicating that they arose through dedifferentiation) (H, arrow). However, under the same conditions, there are no bam-lineage positive GSCs in the bam > puc testis (J), indicating that JNK signaling is required for dedifferentiation. (K) Quantification of the rate of bam-lineage dedifferentiation among GSCs in controls (gray bars) and after repressing the JNK pathway with either UAS-puc (blue bars) or a dominant negative form of basket (UAS-bskK53R) (purple bars). Both transgenes are able to block dedifferentiation, mimicking the effects of bam mis-expression. Scale bars in A-E represent 20 µm, while in G-J they represent 10 µm. Bars in the graphs represent mean ±SE. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.